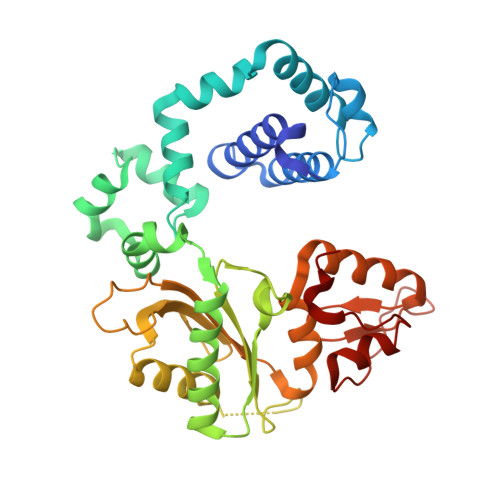
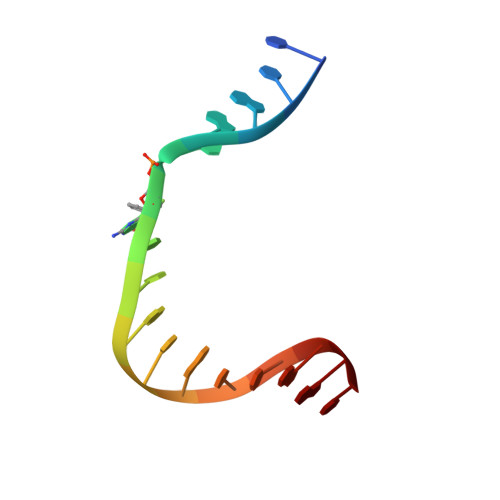
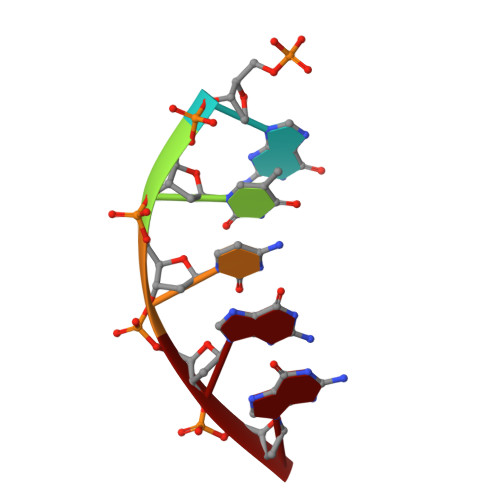
Structural and kinetic studies of the effect of guanine-N7 alkylation and metal cofactors on DNA replication.
Kou, Y., Koag, M.C., Lee, S.(2018) Biochemistry
- PubMed: 29957995
- DOI: https://doi.org/10.1021/acs.biochem.8b00331
- Primary Citation of Related Structures:
4YMN, 4YMO, 4YN4, 5EOZ - PubMed Abstract:
A wide variety of endogenous and exogenous alkylating agents attack DNA to preferentially generate N7-alkylguanine (N7-alkylG) adducts. Studies of the effect of N7-alkylG lesions on biological processes have been difficult in part because of complications arising from the chemical lability of the positively charged N7-alkylG, which can readily produce secondary lesions. To assess the effect of bulky N7-alkylG on DNA replication, we prepared chemically stable N7-benzylguanine (N7bnG)-containing DNA and evaluated nucleotide incorporation opposite the lesion by human DNA polymerase β (polβ), a model enzyme for high-fidelity DNA polymerases. Kinetic studies showed that the N7-benzyl-G lesion greatly inhibited dCTP incorporation by polβ. The crystal structure of polβ incorporating dCTP opposite N7bnG showed a Watson-Crick N7bnG:dCTP structure. The polβ-N7bnG:dCTP structure showed an open protein conformation, a relatively disordered dCTP, and a lack of catalytic metal, which explained the inefficient nucleotide incorporation opposite N7bnG. This indicates that polβ is sensitive to major groove adducts in the templating base side and deters nucleotide incorporation opposite bulky N7-alkylG adducts by adopting a catalytically incompetent conformation. Substituting Mg 2+ for Mn 2+ induced an open-to-closed conformational change due to the presence of catalytic metal and stably bound dCTP and increased the catalytic efficiency by ∼10-fold, highlighting the effect of binding of the incoming nucleotide and catalytic metal on protein conformation and nucleotidyl transfer reaction. Overall, these results suggest that, although bulky alkyl groups at guanine-N7 may not alter base pairing properties of guanine, the major groove-positioned lesions in the template could impede nucleotidyl transfer by some DNA polymerases.
- Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy , The University of Texas at Austin , Austin , Texas 78712 , United States.
Organizational Affiliation: