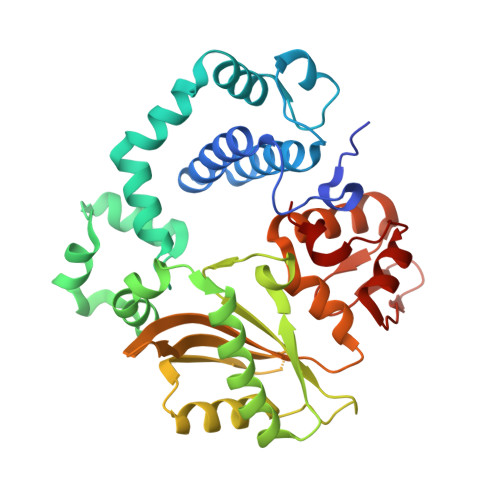
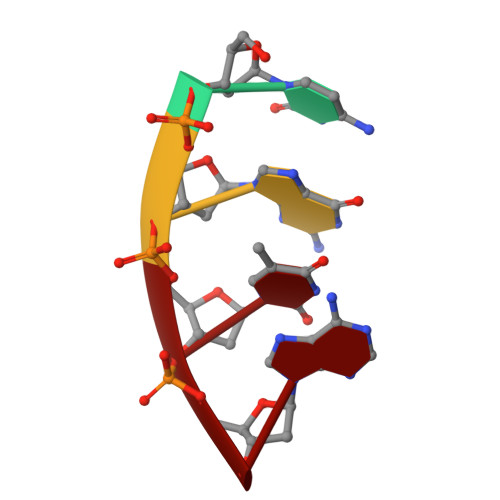
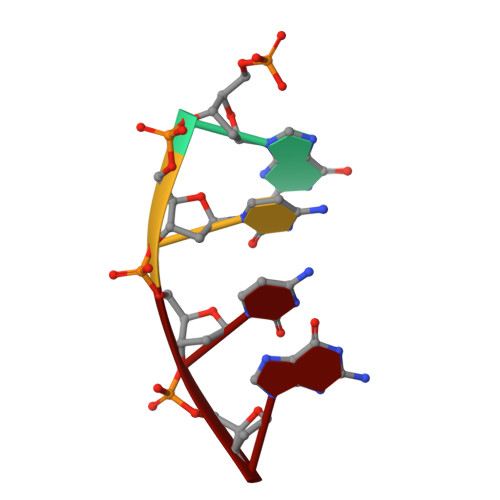
Creative template-dependent synthesis by human polymerase mu.
Moon, A.F., Gosavi, R.A., Kunkel, T.A., Pedersen, L.C., Bebenek, K.(2015) Proc Natl Acad Sci U S A 112: E4530-E4536
- PubMed: 26240373
- DOI: https://doi.org/10.1073/pnas.1505798112
- Primary Citation of Related Structures:
4YCX, 4YD1, 4YD2 - PubMed Abstract:
Among the many proteins used to repair DNA double-strand breaks by nonhomologous end joining (NHEJ) are two related family X DNA polymerases, Pol λ and Pol µ. Which of these two polymerases is preferentially used for filling DNA gaps during NHEJ partly depends on sequence complementarity at the break, with Pol λ and Pol µ repairing complementary and noncomplementary ends, respectively. To better understand these substrate preferences, we present crystal structures of Pol µ on a 2-nt gapped DNA substrate, representing three steps of the catalytic cycle. In striking contrast to Pol λ, Pol µ "skips" the first available template nucleotide, instead using the template base at the 5' end of the gap to direct nucleotide binding and incorporation. This remarkable divergence from canonical 3'-end gap filling is consistent with data on end-joining substrate specificity in cells, and provides insights into polymerase substrate choices during NHEJ.
- Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709.
Organizational Affiliation: