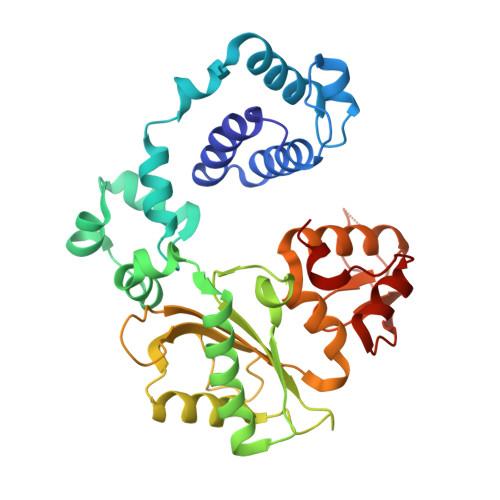
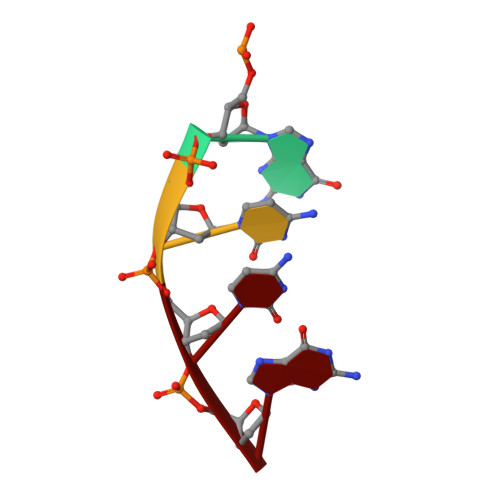
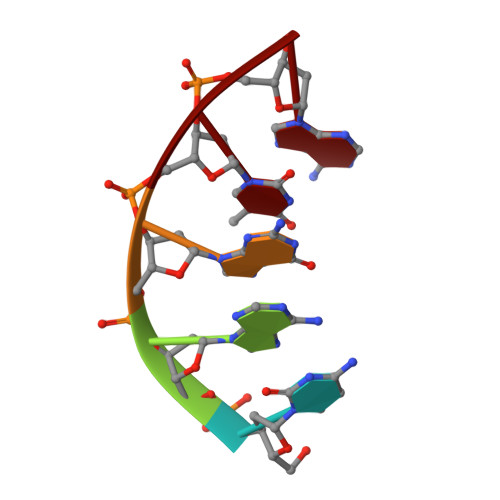
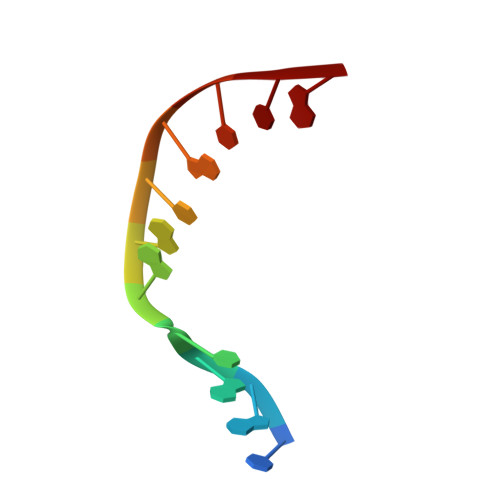
Nucleotide binding interactions modulate dNTP selectivity and facilitate 8-oxo-dGTP incorporation by DNA polymerase lambda.
Burak, M.J., Guja, K.E., Garcia-Diaz, M.(2015) Nucleic Acids Res 43: 8089-8099
- PubMed: 26220180
- DOI: https://doi.org/10.1093/nar/gkv760
- Primary Citation of Related Structures:
4X5V, 4XA5, 4XUS - PubMed Abstract:
8-Oxo-7,8,-dihydro-2'-deoxyguanosine triphosphate (8-oxo-dGTP) is a major product of oxidative damage in the nucleotide pool. It is capable of mispairing with adenosine (dA), resulting in futile, mutagenic cycles of base excision repair. Therefore, it is critical that DNA polymerases discriminate against 8-oxo-dGTP at the insertion step. Because of its roles in oxidative DNA damage repair and non-homologous end joining, DNA polymerase lambda (Pol λ) may frequently encounter 8-oxo-dGTP. Here, we have studied the mechanisms of 8-oxo-dGMP incorporation and discrimination by Pol λ. We have solved high resolution crystal structures showing how Pol λ accommodates 8-oxo-dGTP in its active site. The structures indicate that when mispaired with dA, the oxidized nucleotide assumes the mutagenic syn-conformation, and is stabilized by multiple interactions. Steady-state kinetics reveal that two residues lining the dNTP binding pocket, Ala(510) and Asn(513), play differential roles in dNTP selectivity. Specifically, Ala(510) and Asn(513) facilitate incorporation of 8-oxo-dGMP opposite dA and dC, respectively. These residues also modulate the balance between purine and pyrimidine incorporation. Our results shed light on the mechanisms controlling 8-oxo-dGMP incorporation in Pol λ and on the importance of interactions with the incoming dNTP to determine selectivity in family X DNA polymerases.