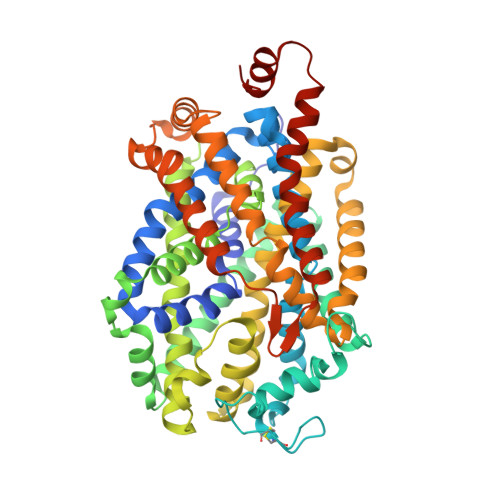
Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Wang, K.H., Penmatsa, A., Gouaux, E.(2015) Nature 521: 322-327
- PubMed: 25970245
- DOI: https://doi.org/10.1038/nature14431
- Primary Citation of Related Structures:
4XP1, 4XP4, 4XP5, 4XP6, 4XP9, 4XPA, 4XPB, 4XPF, 4XPG, 4XPH, 4XPT - PubMed Abstract:
Na(+)/Cl(-)-coupled biogenic amine transporters are the primary targets of therapeutic and abused drugs, ranging from antidepressants to the psychostimulants cocaine and amphetamines, and to their cognate substrates. Here we determine X-ray crystal structures of the Drosophila melanogaster dopamine transporter (dDAT) bound to its substrate dopamine, a substrate analogue 3,4-dichlorophenethylamine, the psychostimulants d-amphetamine and methamphetamine, or to cocaine and cocaine analogues. All ligands bind to the central binding site, located approximately halfway across the membrane bilayer, in close proximity to bound sodium and chloride ions. The central binding site recognizes three chemically distinct classes of ligands via conformational changes that accommodate varying sizes and shapes, thus illustrating molecular principles that distinguish substrates from inhibitors in biogenic amine transporters.
- Vollum Institute, Oregon Health &Science University, 3181 SW Sam Jackson Park Road, Portland, Oregon 97239, USA.
Organizational Affiliation: