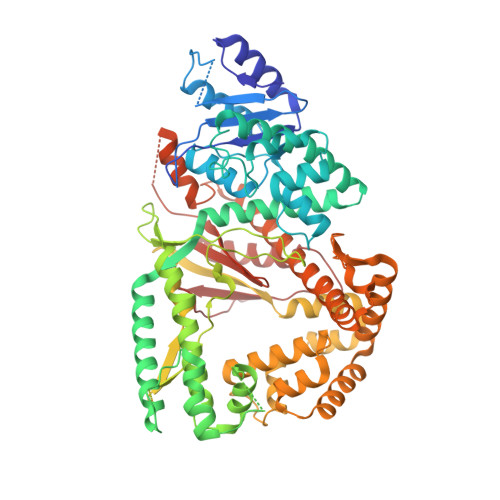
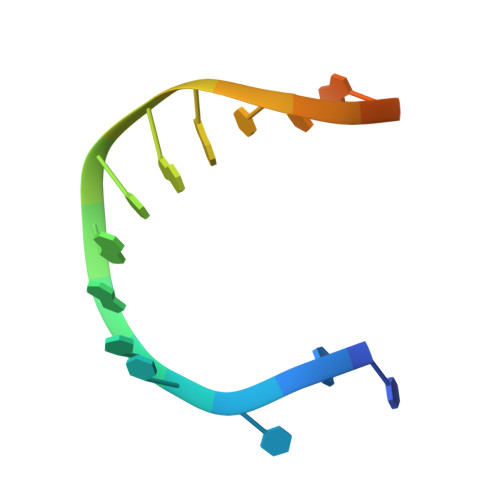
Human DNA polymerase theta grasps the primer terminus to mediate DNA repair.
Zahn, K.E., Averill, A.M., Aller, P., Wood, R.D., Doublie, S.(2015) Nat Struct Mol Biol 22: 304-311
- PubMed: 25775267
- DOI: https://doi.org/10.1038/nsmb.2993
- Primary Citation of Related Structures:
4X0P, 4X0Q - PubMed Abstract:
DNA polymerase θ protects against genomic instability via an alternative end-joining repair pathway for DNA double-strand breaks. Polymerase θ is overexpressed in breast, lung and oral cancers, and reduction of its activity in mammalian cells increases sensitivity to double-strand break-inducing agents, including ionizing radiation. Reported here are crystal structures of the C-terminal polymerase domain from human polymerase θ, illustrating two potential modes of dimerization. One structure depicts insertion of ddATP opposite an abasic-site analog during translesion DNA synthesis. The second structure describes a cognate ddGTP complex. Polymerase θ uses a specialized thumb subdomain to establish unique upstream contacts to the primer DNA strand, including an interaction with the 3'-terminal phosphate from one of five distinctive insertion loops. These observations demonstrate how polymerase θ grasps the primer to bypass DNA lesions or extend poorly annealed DNA termini to mediate end-joining.
- Department of Microbiology and Molecular Genetics, University of Vermont, Burlington, Vermont, USA.
Organizational Affiliation: