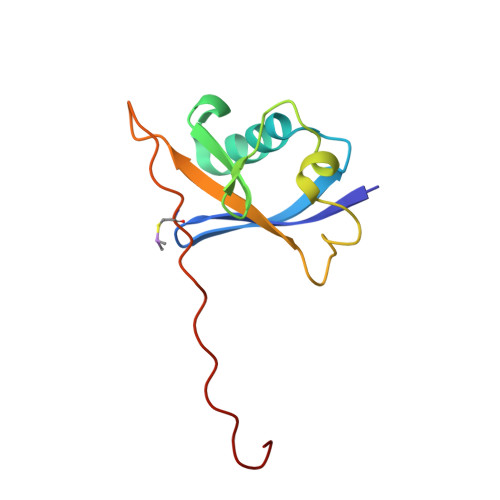
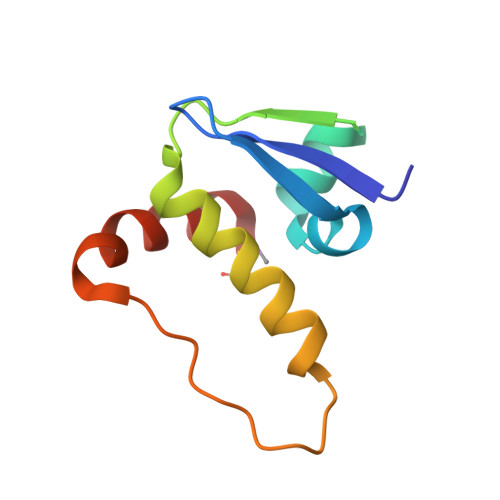
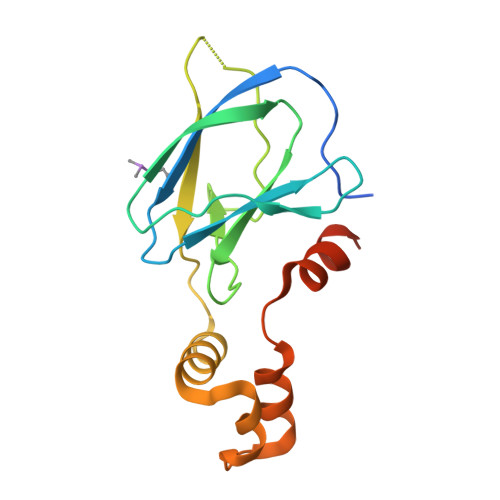
Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
Galdeano, C., Gadd, M.S., Soares, P., Scaffidi, S., Van Molle, I., Birced, I., Hewitt, S., Dias, D.M., Ciulli, A.(2014) J Med Chem 57: 8657-8663
- PubMed: 25166285
- DOI: https://doi.org/10.1021/jm5011258
- Primary Citation of Related Structures:
4W9C, 4W9D, 4W9E, 4W9F, 4W9G, 4W9H, 4W9I, 4W9J, 4W9K, 4W9L - PubMed Abstract:
E3 ubiquitin ligases are attractive targets in the ubiquitin-proteasome system, however, the development of small-molecule ligands has been rewarded with limited success. The von Hippel-Lindau protein (pVHL) is the substrate recognition subunit of the VHL E3 ligase that targets HIF-1α for degradation. We recently reported inhibitors of the pVHL:HIF-1α interaction, however they exhibited moderate potency. Herein, we report the design and optimization, guided by X-ray crystal structures, of a ligand series with nanomolar binding affinities.
- Division of Biological Chemistry and Drug Discovery, College of Life Sciences, University of Dundee , Dow Street, Dundee, DD1 5EH, Scotland, U.K.
Organizational Affiliation: