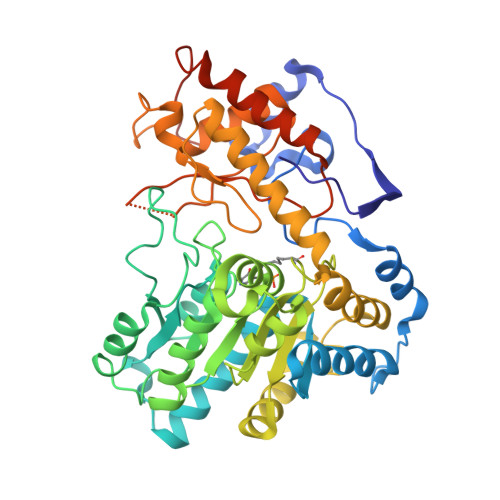
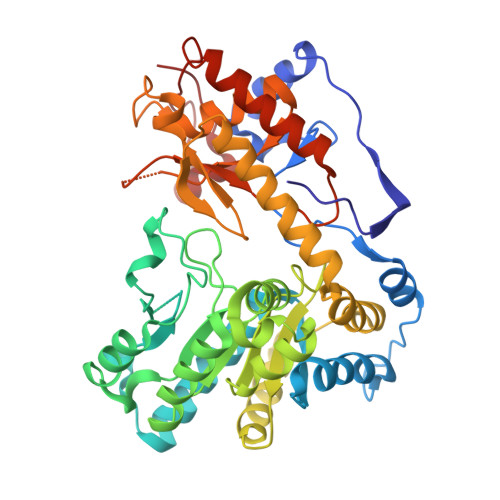
Structures of Escherichia coli tryptophanase in holo and `semi-holo' forms.
Kogan, A., Raznov, L., Gdalevsky, G.Y., Cohen-Luria, R., Almog, O., Parola, A.H., Goldgur, Y.(2015) Acta Crystallogr F Struct Biol Commun 71: 286-290
- PubMed: 25760702
- DOI: https://doi.org/10.1107/S2053230X15000850
- Primary Citation of Related Structures:
4W1Y, 4W4H - PubMed Abstract:
Two crystal forms of Escherichia coli tryptophanase (tryptophan indole-lyase, Trpase) were obtained under the same crystallization conditions. Both forms belonged to the same space group P43212 but had slightly different unit-cell parameters. The holo crystal form, with pyridoxal phosphate (PLP) bound to Lys270 of both polypeptide chains in the asymmetric unit, diffracted to 2.9 Å resolution. The second crystal form diffracted to 3.2 Å resolution. Of the two subunits in the asymmetric unit, one was found in the holo form, while the other appeared to be in the apo form in a wide-open conformation with two sulfate ions bound in the vicinity of the active site. The conformation of all holo subunits is the same in both crystal forms. The structures suggest that Trpase is flexible in the apo form. Its conformation partially closes upon binding of PLP. The closed conformation might correspond to the enzyme in its active state with both cofactor and substrate bound in a similar way as in tyrosine phenol-lyase.
- Department of Chemistry, Ben Gurion University of the Negev, Beer Sheva 84105, Israel.
Organizational Affiliation: