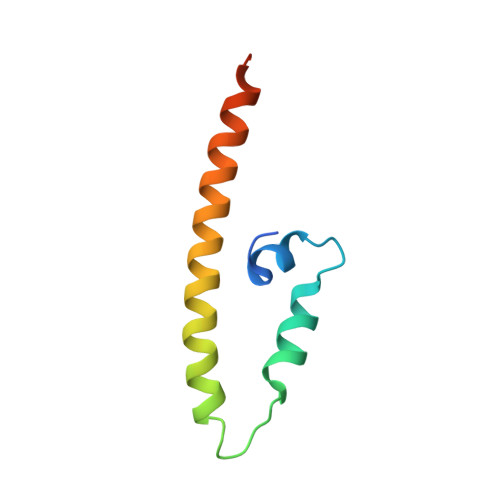
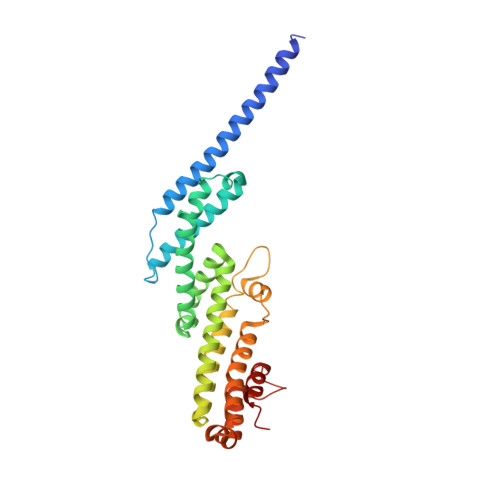
Cog5-Cog7 crystal structure reveals interactions essential for the function of a multisubunit tethering complex.
Ha, J.Y., Pokrovskaya, I.D., Climer, L.K., Shimamura, G.R., Kudlyk, T., Jeffrey, P.D., Lupashin, V.V., Hughson, F.M.(2014) Proc Natl Acad Sci U S A 111: 15762-15767
- PubMed: 25331899
- DOI: https://doi.org/10.1073/pnas.1414829111
- Primary Citation of Related Structures:
4U6U - PubMed Abstract:
The conserved oligomeric Golgi (COG) complex is required, along with SNARE and Sec1/Munc18 (SM) proteins, for vesicle docking and fusion at the Golgi. COG, like other multisubunit tethering complexes (MTCs), is thought to function as a scaffold and/or chaperone to direct the assembly of productive SNARE complexes at the sites of membrane fusion. Reflecting this essential role, mutations in the COG complex can cause congenital disorders of glycosylation. A deeper understanding of COG function and dysfunction will likely depend on elucidating its molecular structure. Despite some progress toward this goal, including EM studies of COG lobe A (subunits 1-4) and higher-resolution structures of portions of Cog2 and Cog4, the structures of COG's eight subunits and the principles governing their assembly are mostly unknown. Here, we report the crystal structure of a complex between two lobe B subunits, Cog5 and Cog7. The structure reveals that Cog5 is a member of the complexes associated with tethering containing helical rods (CATCHR) fold family, with homology to subunits of other MTCs including the Dsl1, exocyst, and Golgi-associated retrograde protein (GARP) complexes. The Cog5-Cog7 interaction is analyzed in relation to the Dsl1 complex, the only other CATCHR-family MTC for which subunit interactions have been characterized in detail. Biochemical and functional studies validate the physiological relevance of the observed Cog5-Cog7 interface, indicate that it is conserved from yeast to humans, and demonstrate that its disruption in human cells causes defects in trafficking and glycosylation.
- Department of Molecular Biology, Princeton University, Princeton, NJ 08544;
Organizational Affiliation: