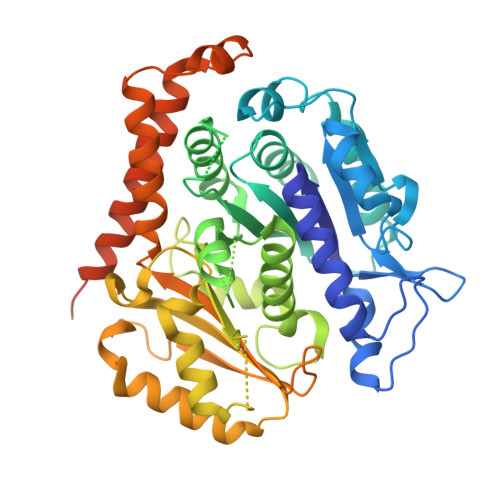
A tethered delivery mechanism explains the catalytic action of a microtubule polymerase.
Ayaz, P., Munyoki, S., Geyer, E.A., Piedra, F.A., Vu, E.S., Bromberg, R., Otwinowski, Z., Grishin, N.V., Brautigam, C.A., Rice, L.M.(2014) Elife 3: e03069-e03069
- PubMed: 25097237
- DOI: https://doi.org/10.7554/eLife.03069
- Primary Citation of Related Structures:
4U3J - PubMed Abstract:
Stu2p/XMAP215 proteins are essential microtubule polymerases that use multiple αβ-tubulin-interacting TOG domains to bind microtubule plus ends and catalyze fast microtubule growth. We report here the structure of the TOG2 domain from Stu2p bound to yeast αβ-tubulin. Like TOG1, TOG2 binds selectively to a fully 'curved' conformation of αβ-tubulin, incompatible with a microtubule lattice. We also show that TOG1-TOG2 binds non-cooperatively to two αβ-tubulins. Preferential interactions between TOGs and fully curved αβ-tubulin that cannot exist elsewhere in the microtubule explain how these polymerases localize to the extreme microtubule end. We propose that these polymerases promote elongation because their linked TOG domains concentrate unpolymerized αβ-tubulin near curved subunits already bound at the microtubule end. This tethering model can explain catalyst-like behavior and also predicts that the polymerase action changes the configuration of the microtubule end.
- Department of Biophysics, UT Southwestern Medical Center, Dallas, United States Department of Biochemistry, UT Southwestern Medical Center, Dallas, United States.
Organizational Affiliation: