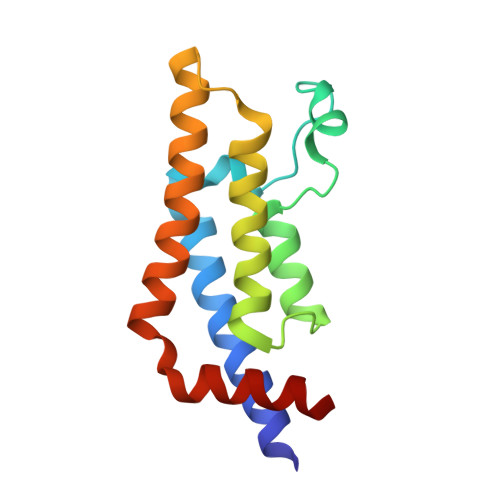
Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Poncet-Montange, G., Zhan, Y., Bardenhagen, J.P., Petrocchi, A., Leo, E., Shi, X., Lee, G.R., Leonard, P.G., Geck Do, M.K., Cardozo, M.G., Andersen, J.N., Palmer, W.S., Jones, P., Ladbury, J.E.(2015) Biochem J 466: 337-346
- PubMed: 25486442
- DOI: https://doi.org/10.1042/BJ20140933
- Primary Citation of Related Structures:
4TT2, 4TT4, 4TT6, 4TTE, 4TU4, 4TU6 - PubMed Abstract:
Preventing histone recognition by bromodomains emerges as an attractive therapeutic approach in cancer. Overexpression of ATAD2 (ATPase family AAA domain-containing 2 isoform A) in cancer cells is associated with poor prognosis making the bromodomain of ATAD2 a promising epigenetic therapeutic target. In the development of an in vitro assay and identification of small molecule ligands, we conducted structure-guided studies which revealed a conformationally flexible ATAD2 bromodomain. Structural studies on apo-, peptide-and small molecule-ATAD2 complexes (by co-crystallization) revealed that the bromodomain adopts a 'closed', histone-compatible conformation and a more 'open' ligand-compatible conformation of the binding site respectively. An unexpected conformational change of the conserved asparagine residue plays an important role in driving the peptide-binding conformation remodelling. We also identified dimethylisoxazole-containing ligands as ATAD2 binders which aided in the validation of the in vitro screen and in the analysis of these conformational studies.
- *Institute for Applied Cancer Science, The University of Texas MD Anderson Cancer Center, Unit 1954, 1515 Holcombe Blvd, Houston, TX 77030, U.S.A.
Organizational Affiliation: