Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Hattori, Y., Kobayashi, K., Deguchi, A., Nohara, Y., Akiyama, T., Teruya, K., Sanjoh, A., Nakagawa, A., Yamashita, E., Akaji, K.(2015) Bioorg Med Chem 23: 5626-5640
- PubMed: 26264846
- DOI: https://doi.org/10.1016/j.bmc.2015.07.023
- Primary Citation of Related Structures:
4TRW, 4TRZ - PubMed Abstract:
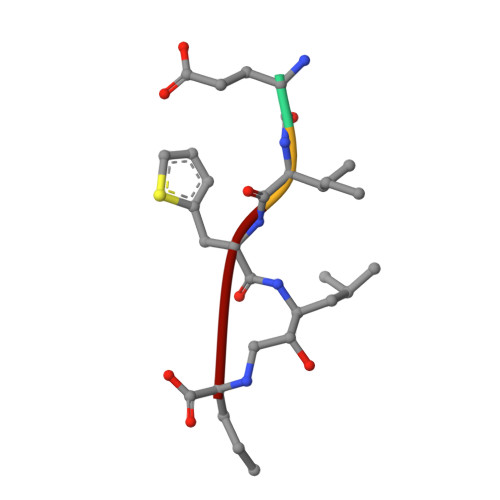
A superior substrate sequence for BACE1 containing transition-state mimics at the scissile site was evaluated as a protease inhibitor. Hydroxymethylcarbonyl (HMC) and hydroxyethylamine (HEA) isosteres were selected as the transition state mimics, and incorporated into the scissile site of the superior sequence covering the P4 to P1' sites (Glu-Ile-Thi-Thi(*)Nva; (*)denotes the cleavage site). Isosteres having different absolute configurations of the hydroxyl group were synthesized separately, and the effect of the configuration was evaluated. Configuration of the hydroxyl group of each isostere showed a marked effect on the inhibitory activity; anti-configuration to the scissile site substituent had potent inhibitory activity in an HMC-type inhibitor, whereas anti-configuration of HEA-type inhibitors showed no inhibitory activity. Structural evaluations based on X-ray crystallographic analyses of recombinant BACE1 in complex with each inhibitor provided insights into the protein-ligand interactions, especially at the prime sites.
- Department of Medicinal Chemistry, Kyoto Pharmaceutical University, Yamashina-ku, Kyoto 607-8412, Japan.
Organizational Affiliation: