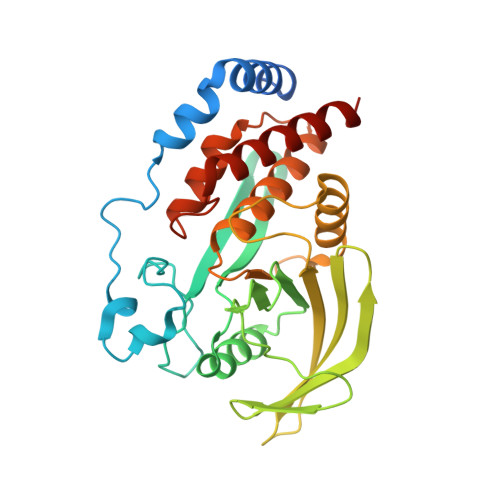
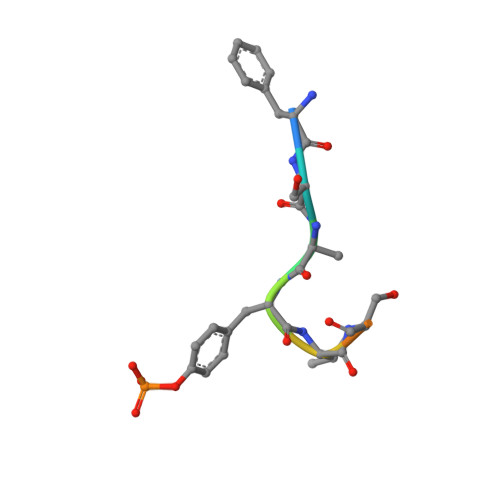
Substrate specificity and plasticity of FERM-containing protein tyrosine phosphatases.
Chen, K.E., Li, M.Y., Chou, C.C., Ho, M.R., Chen, G.C., Meng, T.C., Wang, A.H.(2015) Structure 23: 653-664
- PubMed: 25728925
- DOI: https://doi.org/10.1016/j.str.2015.01.017
- Primary Citation of Related Structures:
4RH5, 4RH9, 4RHG, 4RI4, 4RI5, 4S0G - PubMed Abstract:
Epidermal growth factor receptor (EGFR) pathway substrate 15 (Eps15) is a newly identified substrate for protein tyrosine phosphatase N3 (PTPN3), which belongs to the FERM-containing PTP subfamily comprising five members including PTPN3, N4, N13, N14, and N21. We solved the crystal structures of the PTPN3-Eps15 phosphopeptide complex and found that His812 of PTPN3 and Pro850 of Eps15 are responsible for the specific interaction between them. We defined the critical role of the additional residue Tyr676 of PTPN3, which is replaced by Ile939 in PTPN14, in recognition of tyrosine phosphorylated Eps15. The WPD loop necessary for catalysis is present in all members but not PTPN21. We identified that Glu instead of Asp in the WPE loop contributes to the catalytic incapability of PTPN21 due to an extended distance beyond protonation targeting a phosphotyrosine substrate. Together with in vivo validations, our results provide novel insights into the substrate specificity and plasticity of FERM-containing PTPs.
- Institute of Biological Chemistry, Academia Sinica, Taipei 115, Taiwan.
Organizational Affiliation: