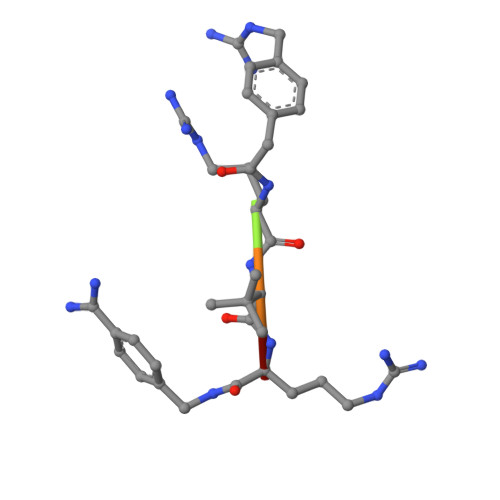
Novel Furin Inhibitors with Potent Anti-infectious Activity.
Hardes, K., Becker, G.L., Lu, Y., Dahms, S.O., Kohler, S., Beyer, W., Sandvig, K., Yamamoto, H., Lindberg, I., Walz, L., von Messling, V., Than, M.E., Garten, W., Steinmetzer, T.(2015) ChemMedChem 10: 1218-1231
- PubMed: 25974265
- DOI: https://doi.org/10.1002/cmdc.201500103
- Primary Citation of Related Structures:
4RYD - PubMed Abstract:
New peptidomimetic furin inhibitors with unnatural amino acid residues in the P3 position were synthesized. The most potent compound 4-guanidinomethyl-phenylacteyl-Arg-Tle-Arg-4-amidinobenzylamide (MI-1148) inhibits furin with a Ki value of 5.5 pM. The derivatives also strongly inhibit PC1/3, whereas PC2 is less affected. Selected inhibitors were tested in cell culture for antibacterial and antiviral activity against infectious agents known to be dependent on furin activity. A significant protective effect against anthrax and diphtheria toxin was observed in the presence of the furin inhibitors. Furthermore, the spread of the highly pathogenic H5N1 and H7N1 avian influenza viruses and propagation of canine distemper virus was strongly inhibited. Inhibitor MI-1148 was crystallized in complex with human furin. Its N-terminal guanidinomethyl group in the para position of the P5 phenyl ring occupies the same position as that found previously for a structurally related inhibitor containing this substitution in the meta position, thereby maintaining all of the important P5 interactions. Our results confirm that the inhibition of furin is a promising strategy for a short-term treatment of acute infectious diseases.
- Institute of Pharmaceutical Chemistry, Philipps University, Marbacher Weg 6, 35032 Marburg (Germany).
Organizational Affiliation: