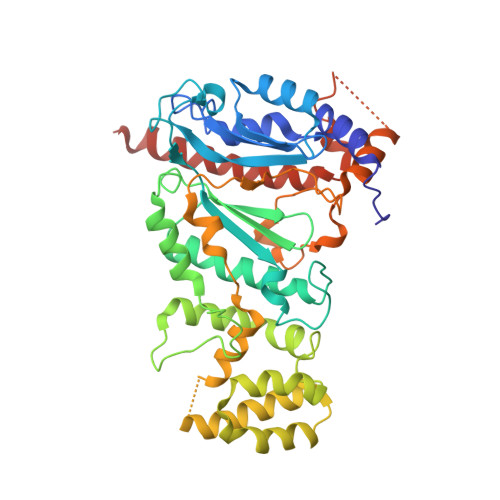
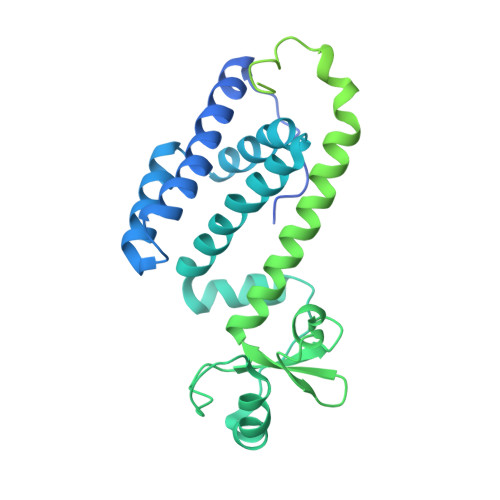
Crystal structure of the human primase.
Baranovskiy, A.G., Zhang, Y., Suwa, Y., Babayeva, N.D., Gu, J., Pavlov, Y.I., Tahirov, T.H.(2015) J Biological Chem 290: 5635-5646
- PubMed: 25550159
- DOI: https://doi.org/10.1074/jbc.M114.624742
- Primary Citation of Related Structures:
4RR2 - PubMed Abstract:
DNA replication in bacteria and eukaryotes requires the activity of DNA primase, a DNA-dependent RNA polymerase that lays short RNA primers for DNA polymerases. Eukaryotic and archaeal primases are heterodimers consisting of small catalytic and large accessory subunits, both of which are necessary for RNA primer synthesis. Understanding of RNA synthesis priming in eukaryotes is currently limited due to the lack of crystal structures of the full-length primase and its complexes with substrates in initiation and elongation states. Here we report the crystal structure of the full-length human primase, revealing the precise overall organization of the enzyme, the relative positions of its functional domains, and the mode of its interaction with modeled DNA and RNA. The structure indicates that the dramatic conformational changes in primase are necessary to accomplish the initiation and then elongation of RNA synthesis. The presence of a long linker between the N- and C-terminal domains of p58 provides the structural basis for the bulk of enzyme's conformational flexibility. Deletion of most of this linker affected the initiation and elongation steps of the primer synthesis.
- From the Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, Nebraska 68198.
Organizational Affiliation: