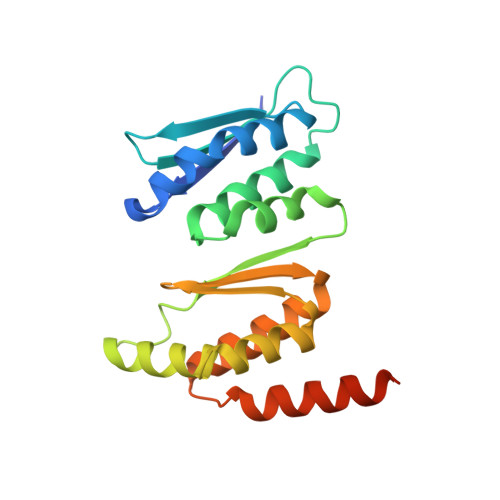
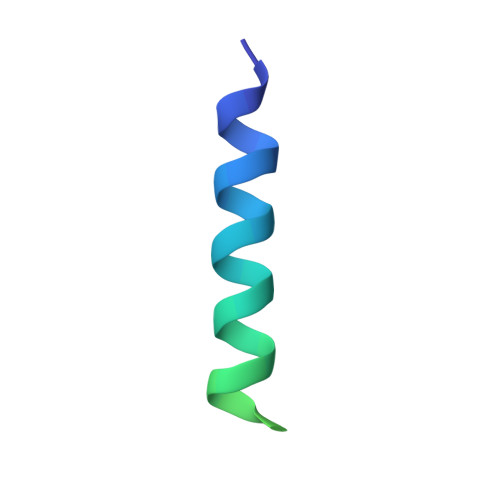
Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast
Zheng, S., Lan, P., Liu, X., Ye, K.(2014) J Biological Chem 289: 22692-22703
- PubMed: 24990943
- DOI: https://doi.org/10.1074/jbc.M114.584490
- Primary Citation of Related Structures:
4QMF - PubMed Abstract:
Ribosome formation in Saccharomyces cerevisiae requires a large number of transiently associated assembly factors that coordinate processing and folding of pre-rRNA and binding of ribosomal proteins. Krr1 and Faf1 are two interacting proteins present in early 90 S precursor particles of the small ribosomal subunit. Here, we determined a co-crystal structure of the core domain of Krr1 bound to a 19-residue fragment of Faf1 at 2.8 Å resolution. The structure reveals that Krr1 consists of two packed K homology (KH) domains, KH1 and KH2, and resembles archaeal Dim2-like proteins. We show that KH1 is a divergent KH domain that lacks the RNA-binding GXXG motif and is involved in binding another assembly factor, Kri1. KH2 contains a canonical RNA-binding surface and additionally associates with an α-helix of Faf1. Specific disruption of the Krr1-Faf1 interaction impaired early 18 S rRNA processing at sites A0, A1, and A2 and caused cell lethality, but it did not prevent incorporation of the two proteins into pre-ribosomes. The Krr1-Faf1 interaction likely maintains a critical conformation of 90 S pre-ribosomes required for pre-rRNA processing. Our results illustrate the versatility of KH domains in protein interaction and provide insight into the role of Krr1-Faf1 interaction in ribosome biogenesis.
- Department of Biochemistry and Molecular Biology, College of Life Sciences, Beijing Normal University, Beijing 100875,; National Institute of Biological Sciences at Beijing, Beijing 102206, and.
Organizational Affiliation: