Orally Active 7-Substituted (4-Benzylphthalazin-1-yl)-2-methylpiperazin-1-yl]nicotinonitriles as Active-Site Inhibitors of Sphingosine 1-Phosphate Lyase for the Treatment of Multiple Sclerosis.
Weiler, S., Braendlin, N., Beerli, C., Bergsdorf, C., Schubart, A., Srinivas, H., Oberhauser, B., Billich, A.(2014) J Med Chem 57: 5074-5084
- PubMed: 24809814
- DOI: https://doi.org/10.1021/jm500338n
- Primary Citation of Related Structures:
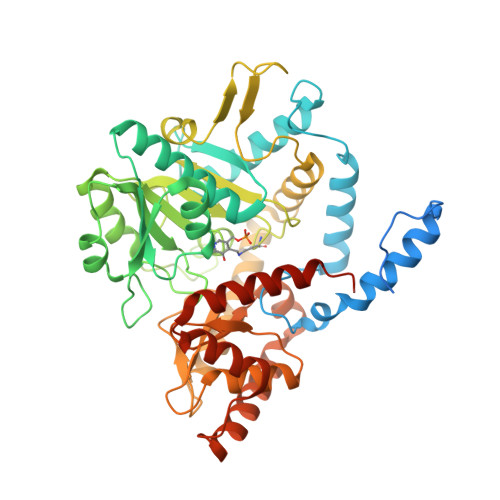
4Q6R - PubMed Abstract:
Sphingosine 1-phosphate (S1P) lyase has recently been implicated as a therapeutic target for the treatment of multiple sclerosis (MS), based on studies in a genetic mouse model. Potent active site directed inhibitors of the enzyme are not known so far. Here we describe the discovery of (4-benzylphthalazin-1-yl)-2-methylpiperazin-1-yl]nicotinonitrile 5 in a high-throughput screen using a biochemical assay, and its further optimization. This class of compounds was found to inhibit catalytic activity of S1PL by binding to the active site of the enzyme, as seen in the cocrystal structure of derivative 31 with the homodimeric human S1P lyase. 31 induces profound reduction of peripheral T cell numbers after oral dosage and confers pronounced protection in a rat model of multiple sclerosis. In conclusion, this novel class of direct S1P lyase inhibitors provides excellent tools to further explore the therapeutic potential of T cell-targeted therapies in multiple sclerosis and other autoimmune and inflammatory diseases.
- Novartis Institutes for BioMedical Research , Basel, CH-4002, Switzerland.
Organizational Affiliation: