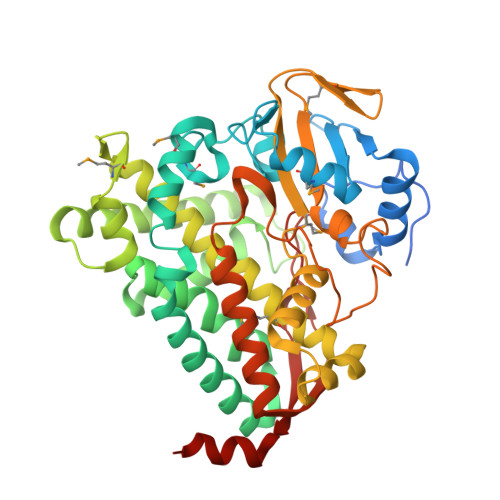
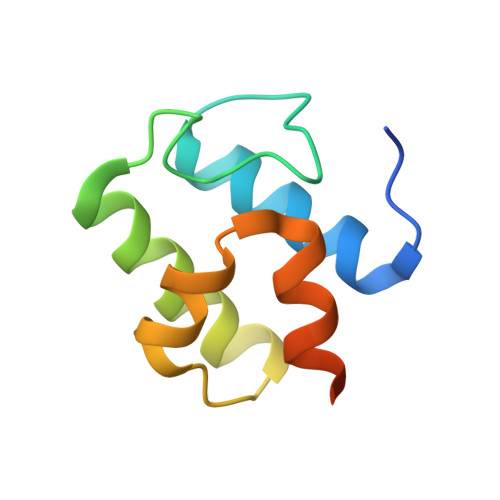
The structure of a transient complex of a nonribosomal Peptide synthetase and a cytochrome p450 monooxygenase.
Haslinger, K., Brieke, C., Uhlmann, S., Sieverling, L., Sussmuth, R.D., Cryle, M.J.(2014) Angew Chem Int Ed Engl 53: 8518-8522
- PubMed: 25044735
- DOI: https://doi.org/10.1002/anie.201404977
- Primary Citation of Related Structures:
4PWV, 4PXH - PubMed Abstract:
Studying the interplay between nonribosomal peptide synthetases (NRPS), a major source of secondary metabolites, and crucial external modifying enzymes is a challenging task since the interactions involved are often transient in nature. By applying a range of synthetic inhibitor-type compounds, a stabilized complex appropriate for structural analysis was generated for such a tailoring enzyme and an NRPS domain. The complex studied comprises an NRPS peptidyl carrier protein (PCP) domain bound to the Cytochrome P450 enzyme that is crucial for the provision of β-hydroxylated amino acid precursors in the biosynthesis of the cyclic depsipeptide skyllamycin. The structure reveals that complex formation is governed by hydrophobic interactions, the presence of which can be controlled through minor alterations in PCP structure that enable selectivity amongst multiple highly similar PCP domains.
- Department of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Jahnstrasse 29, 69120 Heidelberg (Germany).
Organizational Affiliation: