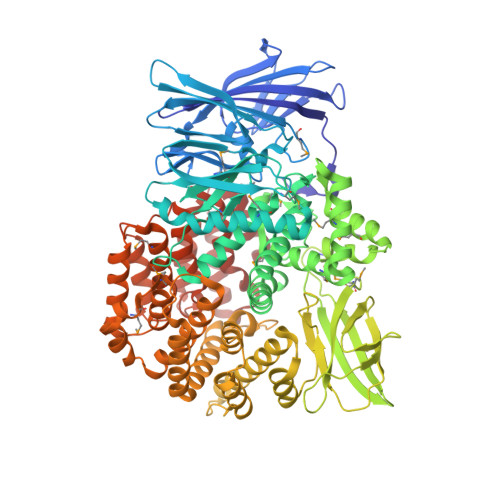
Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine
Nocek, B., Vassiliou, S., Mulligan, R., Weglarz-Tomczak, E., Berlicki, L., Pawelczak, M., Joachimiak, A., Mucha, A., Midwest Center for Structural Genomics (MCSG)To be published.