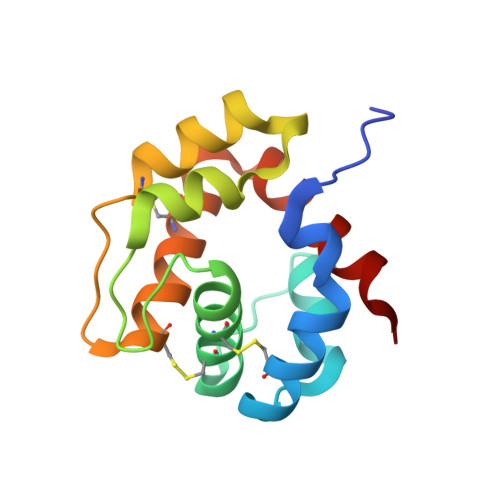
Crystal structure of the Locusta migratoria odorant binding protein.
Zheng, J., Li, J., Han, L., Wang, Y., Wu, W., Qi, X., Tao, Y., Zhang, L., Zhang, Z., Chen, Z.(2015) Biochem Biophys Res Commun 456: 737-742
- PubMed: 25522876
- DOI: https://doi.org/10.1016/j.bbrc.2014.12.048
- Primary Citation of Related Structures:
4PT1 - PubMed Abstract:
Locusta migratoria (Lmig) causes enormous losses to agricultural products, especially because it often infests the world with great swarms as locust plagues. Locusts find their plant hosts on which they feed through their olfactory system, in which odorant binding proteins (OBPs) play an important role. Previous study indicated that the amino acid sequences of LmigOBP showed low similarity to OBPs from other insect orders and we speculated that it might perform unique binding behavior. Here, we solved the first LmigOBP1 structure at 1.65Å, which is a monomer in solution and disulfide bonds play a key role in maintaining its function. We show that LmigOBP1 possesses a unique seventh α-helix, which is located at the surface with strong interactions with the LmigOBP1 scaffold consisting of other six α-helices. Moreover, the seventh α-helix forms a wall of an "L" shaped internal hydrophobic cavity to accommodate linear ligands, which is consistent with the binding experiments. We also demonstrate that the ligand-binding pocket in LmigOBP1 is greatly different from that in the closest homologs mosquito OBPs. Taken together, this study provides a structural basis for designing small inhibitors to control locust.
- State Key Laboratory of Agrobiotechnology, China Agricultural University, Beijing 100193, China.
Organizational Affiliation: