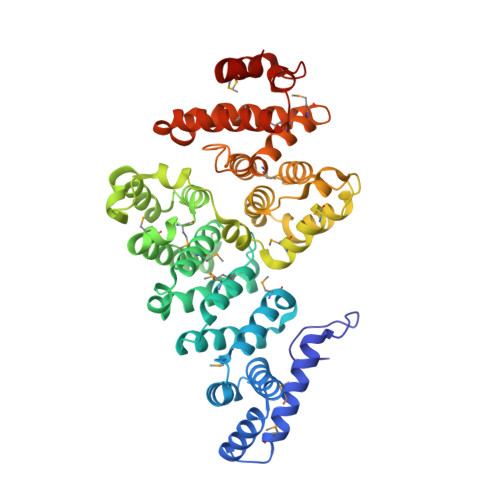
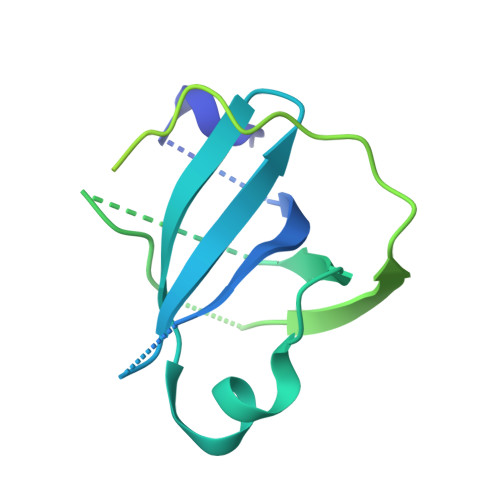
Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Ren, W., Chen, H., Sun, Q., Tang, X., Lim, S.C., Huang, J., Song, H.(2014) Cell Rep 6: 982-991
- PubMed: 24630995
- DOI: https://doi.org/10.1016/j.celrep.2014.02.020
- Primary Citation of Related Structures:
4OWT, 4OWW, 4OWX - PubMed Abstract:
The SOSS1 complex comprising SOSSA, SOSSB1, and SOSSC senses single-stranded DNA (ssDNA) and promotes repair of DNA double-strand breaks (DSBs). But how SOSS1 is assembled and recognizes ssDNA remains elusive. The crystal structure of the N-terminal half of SOSSA (SOSSAN) in complex with SOSSB1 and SOSSC showed that SOSSAN serves as a scaffold to bind both SOSSB1 and SOSSC for assembly of the SOSS1 complex. The structures of SOSSAN/B1 in complex with a 12 nt ssDNA and SOSSAN/B1/C in complex with a 35 nt ssDNA showed that SOSSB1 interacts with both SOSSAN and ssDNA via two distinct surfaces. Recognition of ssDNA with a length of up to nine nucleotides is mediated solely by SOSSB1, whereas neither SOSSC nor SOSSAN are critical for ssDNA binding. These results reveal the structural basis of SOSS1 assembly and provide a framework for further study of the mechanism governing longer ssDNA recognition by the SOSS1 complex during DSB repair.
- Life Sciences Institute, Zhejiang University, 388 Yuhangtang Road, Hangzhou 310058, China; Institute of Molecular and Cell Biology, 61 Biopolis Drive, Singapore 138673, Singapore.
Organizational Affiliation: