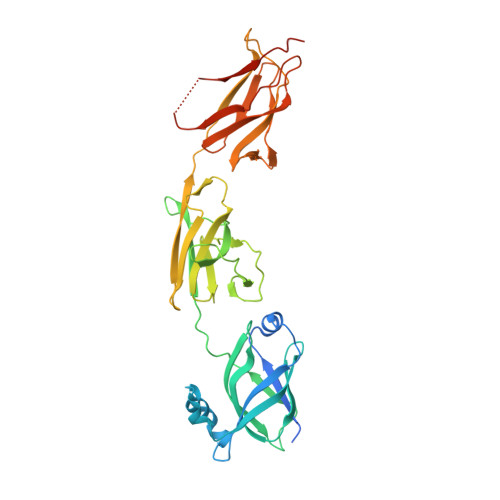
Structural Basis of Pilus Anchoring by the Ancillary Pilin RrgC of Streptococcus pneumoniae.
Shaik, M.M., Maccagni, A., Tourcier, G., Di Guilmi, A.M., Dessen, A.(2014) J Biological Chem 289: 16988-16997
- PubMed: 24755220
- DOI: https://doi.org/10.1074/jbc.M114.555854
- Primary Citation of Related Structures:
4OQ1 - PubMed Abstract:
Pili are surface-attached, fibrous virulence factors that play key roles in the pathogenesis process of a number of bacterial agents. Streptococcus pneumoniae is a causative agent of pneumonia and meningitis, and the appearance of drug-resistance organisms has made its treatment challenging, especially in developing countries. Pneumococcus-expressed pili are composed of three structural proteins: RrgB, which forms the polymerized backbone, RrgA, the tip-associated adhesin, and RrgC, which presumably associates the pilus with the bacterial cell wall. Despite the fact that the structures of both RrgA and RrgB were known previously, structural information for RrgC was still lacking, impeding the analysis of a complete model of pilus architecture. Here, we report the structure of RrgC to 1.85 Å and reveal that it is a three-domain molecule stabilized by two intradomain isopeptide bonds. RrgC does not depend on pilus-specific sortases to become attached to the cell wall; instead, it binds the preformed pilus to the peptidoglycan by employing the catalytic activity of SrtA. A comprehensive model of the type 1 pilus from S. pneumoniae is also presented.
- From the Institut de Biologie Structurale (IBS), Université Grenoble Alpes, 41 avenue des Martyrs, 38044 Grenoble, France, the Commissariat à l'Energie Atomique (CEA), 38000 Grenoble, France, the Centre National de la Recherche Scientifique (CNRS), UMR 5075, Grenoble, France, and.
Organizational Affiliation: