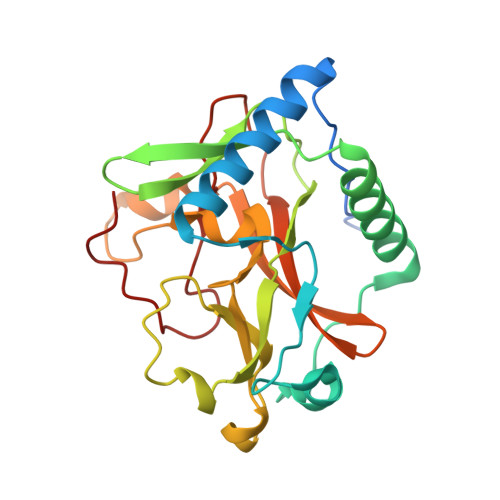
Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication.
Jacob, Y., Bergamin, E., Donoghue, M.T., Mongeon, V., LeBlanc, C., Voigt, P., Underwood, C.J., Brunzelle, J.S., Michaels, S.D., Reinberg, D., Couture, J.F., Martienssen, R.A.(2014) Science 343: 1249-1253
- PubMed: 24626927
- DOI: https://doi.org/10.1126/science.1248357
- Primary Citation of Related Structures:
4O30 - PubMed Abstract:
Histone variants have been proposed to act as determinants for posttranslational modifications with widespread regulatory functions. We identify a histone-modifying enzyme that selectively methylates the replication-dependent histone H3 variant H3.1. The crystal structure of the SET domain of the histone H3 lysine-27 (H3K27) methyltransferase ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) in complex with a H3.1 peptide shows that ATXR5 contains a bipartite catalytic domain that specifically "reads" alanine-31 of H3.1. Variation at position 31 between H3.1 and replication-independent H3.3 is conserved in plants and animals, and threonine-31 in H3.3 is responsible for inhibiting the activity of ATXR5 and its paralog, ATXR6. Our results suggest a simple model for the mitotic inheritance of the heterochromatic mark H3K27me1 and the protection of H3.3-enriched genes against heterochromatization during DNA replication.
- Howard Hughes Medical Institute-Gordon and Betty Moore Foundation, Watson School of Biological Sciences, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: