Insight into Substrate Binding Mechanism in Thymidylate Kinase based on Intrinsic Dynamics of the Active Site
Biswas, A., Jeyakanthan, J., Sekar, K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable thymidylate kinase | 213 | Sulfurisphaera tokodaii str. 7 | Mutation(s): 0 Gene Names: STK_15430, tmk, tmk STK_15430 EC: 2.7.4.9 | 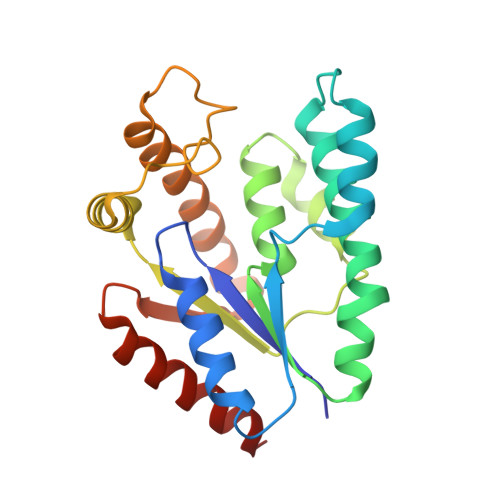 | |
UniProt | |||||
Find proteins for Q970Q8 (Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7)) Explore Q970Q8 Go to UniProtKB: Q970Q8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q970Q8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | C [auth A] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| 1PE Query on 1PE | X [auth B] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| PG4 Query on PG4 | N [auth B] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | O [auth B] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | E [auth A] F [auth A] G [auth A] H [auth A] I [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| PO4 Query on PO4 | D [auth A], P [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| EDO Query on EDO | J [auth A] K [auth A] L [auth A] M [auth A] T [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 49.621 | α = 90 |
| b = 62.643 | β = 90 |
| c = 138.161 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |