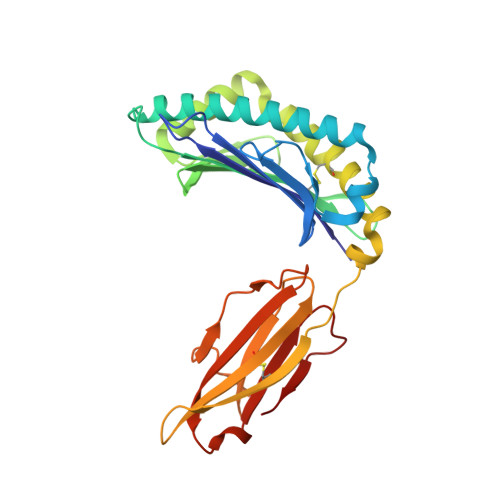
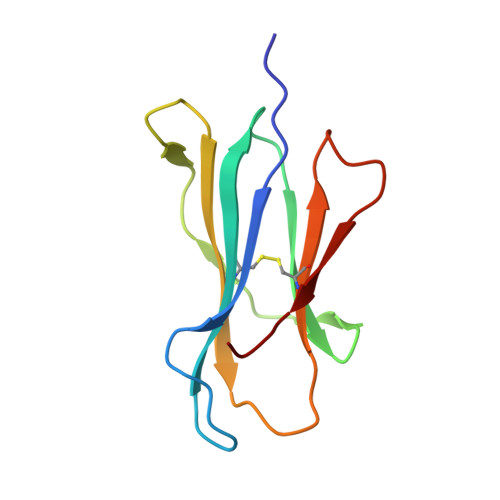
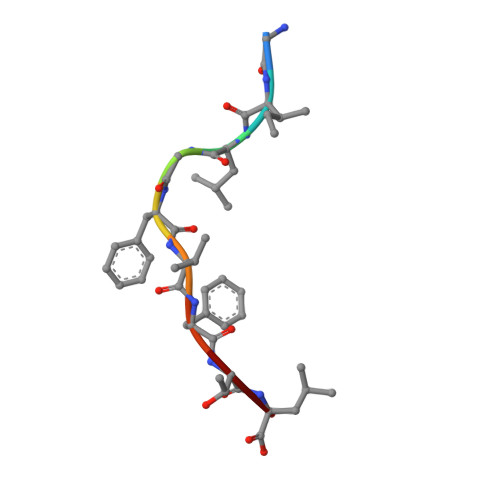
The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans.
Choo, J.A., Liu, J., Toh, X., Grotenbreg, G.M., Ren, E.C.(2014) J Virol 88: 10613-10623
- PubMed: 24990997
- DOI: https://doi.org/10.1128/JVI.00855-14
- Primary Citation of Related Structures:
4NT6 - PubMed Abstract:
Cytotoxic T lymphocytes recognizing conserved peptide epitopes are crucial for protection against influenza A virus (IAV) infection. The CD8 T cell response against the M158-66 (GILGFVFTL) matrix protein epitope is immunodominant when restricted by HLA-A*02, a major histocompatibility complex (MHC) molecule expressed by approximately half of the human population. Here we report that the GILGFVFTL peptide is restricted by multiple HLA-C*08 alleles as well. We observed that M158-66 was able to elicit cytotoxic T lymphocyte (CTL) responses in both HLA-A*02- and HLA-C*08-positive individuals and that GILGFVFTL-specific CTLs in individuals expressing both restriction elements were distinct and not cross-reactive. The crystal structure of GILGFVFTL-HLA-C*08:01 was solved at 1.84 Å, and comparison with the known GILGFVFTL-HLA-A*02:01 structure revealed that the antigen bound both complexes in near-identical conformations, accommodated by binding pockets shaped from shared as well as unique residues. This discovery of degenerate peptide presentation by both HLA-A and HLA-C allelic variants eliciting unique CTL responses to IAV infection contributes fundamental knowledge with important implications for vaccine development strategies. The presentation of influenza A virus peptides to elicit immunity is thought to be narrowly restricted, with a single peptide presented by a specific HLA molecule. In this study, we show that the same influenza A virus peptide can be more broadly presented by both HLA-A and HLA-C molecules. This discovery may help to explain the differences in immunity to influenza A virus between individuals and populations and may also aid in the design of vaccines.
- Department of Microbiology, National University of Singapore, Singapore Immunology Programme, National University of Singapore, Singapore.
Organizational Affiliation: