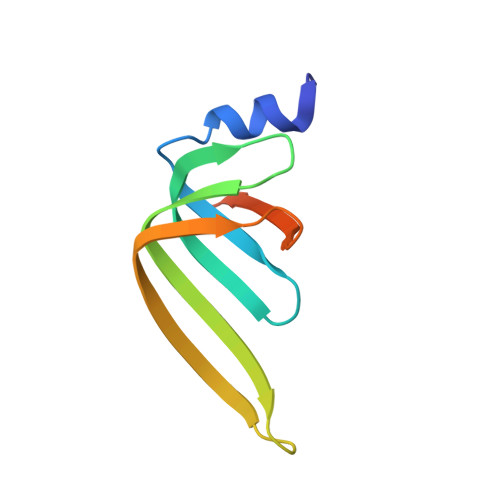
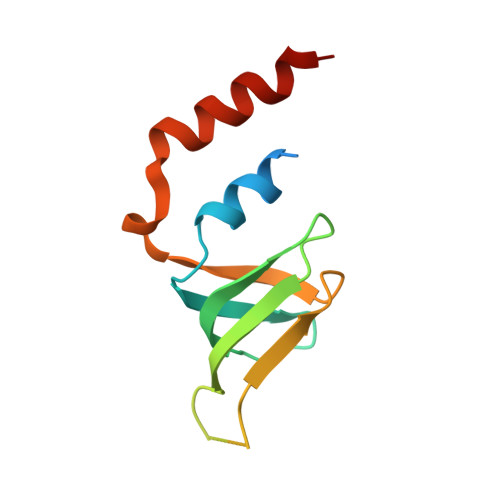
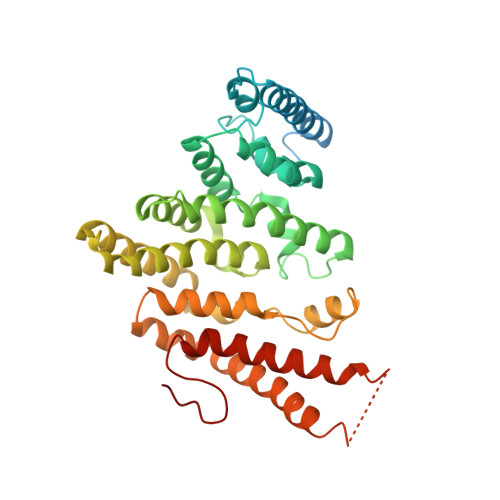
Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation
Wu, D., Muhlrad, D., Bowler, M.W., Jiang, S., Liu, Z., Parker, R., Song, H.(2014) Cell Res 24: 233-246
- PubMed: 24247251
- DOI: https://doi.org/10.1038/cr.2013.152
- Primary Citation of Related Structures:
4N0A - PubMed Abstract:
The evolutionarily conserved Lsm1-7-Pat1 complex is the most critical activator of mRNA decapping in eukaryotic cells and plays many roles in normal decay, AU-rich element-mediated decay, and miRNA silencing, yet how Pat1 interacts with the Lsm1-7 complex is unknown. Here, we show that Lsm2 and Lsm3 bridge the interaction between the C-terminus of Pat1 (Pat1C) and the Lsm1-7 complex. The Lsm2-3-Pat1C complex and the Lsm1-7-Pat1C complex stimulate decapping in vitro to a similar extent and exhibit similar RNA-binding preference. The crystal structure of the Lsm2-3-Pat1C complex shows that Pat1C binds to Lsm2-3 to form an asymmetric complex with three Pat1C molecules surrounding a heptameric ring formed by Lsm2-3. Structure-based mutagenesis revealed the importance of Lsm2-3-Pat1C interactions in decapping activation in vivo. Based on the structure of Lsm2-3-Pat1C, a model of Lsm1-7-Pat1 complex is constructed and how RNA binds to this complex is discussed.
- Institute of Molecular and Cell Biology, 61 Biopolis Drive, Singapore 138673.
Organizational Affiliation: