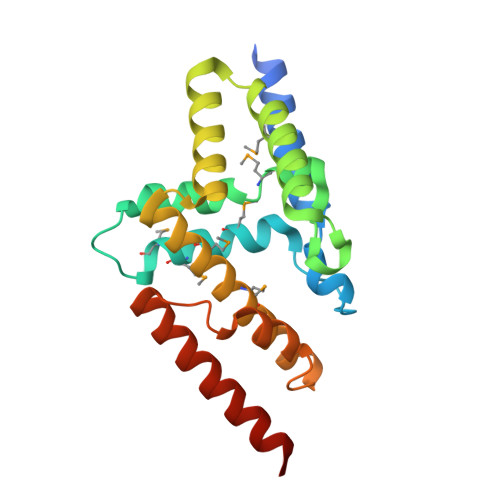
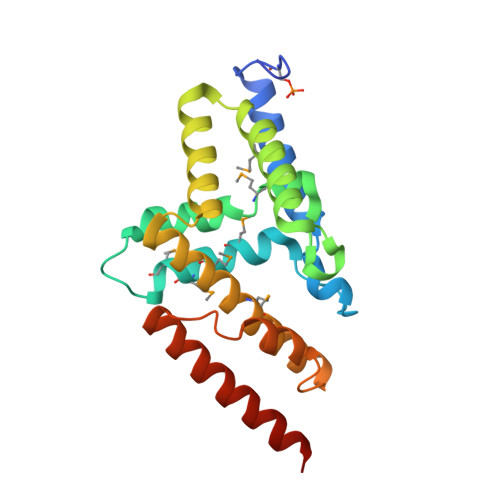
Three dimensional structure of mutant D143A of human HD domain-containing protein 2, Northeast Structural Genomics Consortium (NESG) Target HR6723
Kuzin, A., Su, M., Yakunin, A., Beloglazova, O., Seetharaman, J., Maglaqui, M., Xiao, R., Lee, D., Brown, G., Flick, R., Everett, J.K., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.