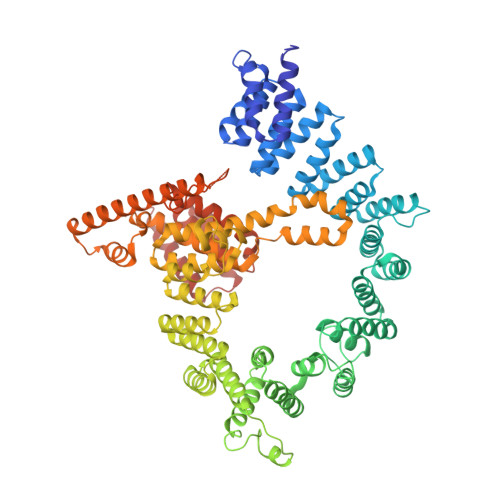
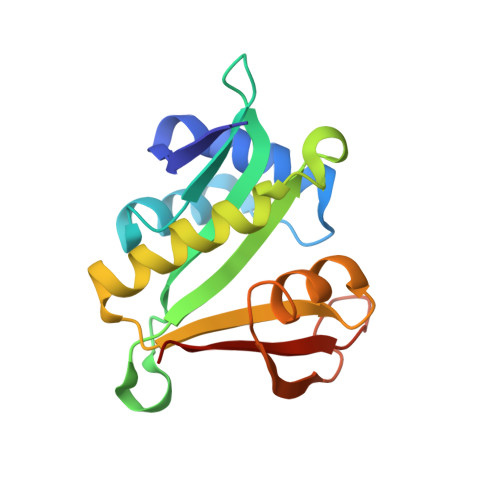
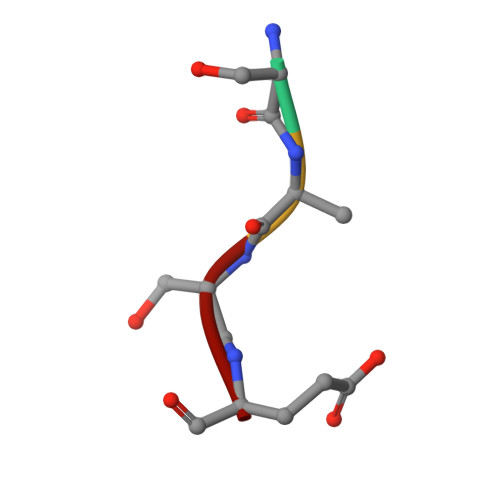
Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.
Liszczak, G., Goldberg, J.M., Foyn, H., Petersson, E.J., Arnesen, T., Marmorstein, R.(2013) Nat Struct Mol Biol 20: 1098-1105
- PubMed: 23912279
- DOI: https://doi.org/10.1038/nsmb.2636
- Primary Citation of Related Structures:
4KVM, 4KVO, 4KVX - PubMed Abstract:
N-terminal acetylation is ubiquitous among eukaryotic proteins and controls a myriad of biological processes. Of the N-terminal acetyltransferases (NATs) that facilitate this cotranslational modification, the heterodimeric NatA complex has the most diversity for substrate selection and modifies the majority of all N-terminally acetylated proteins. Here, we report the X-ray crystal structure of the 100-kDa holo-NatA complex from Schizosaccharomyces pombe, in the absence and presence of a bisubstrate peptide-CoA-conjugate inhibitor, as well as the structure of the uncomplexed Naa10p catalytic subunit. The NatA-Naa15p auxiliary subunit contains 13 tetratricopeptide motifs and adopts a ring-like topology that wraps around the NatA-Naa10p subunit, an interaction that alters the Naa10p active site for substrate-specific acetylation. These studies have implications for understanding the mechanistic details of other NAT complexes and how regulatory subunits modulate the activity of the broader family of protein acetyltransferases.
- 1] Program in Gene Expression and Regulation, Wistar Institute, Philadelphia, Pennsylvania, USA. [2] Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Organizational Affiliation: