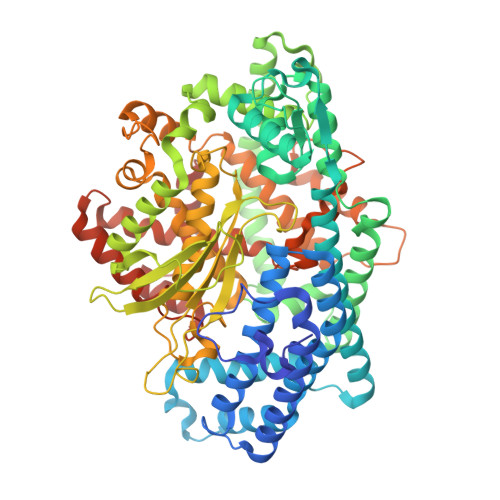
Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts.
Kmiec, B., Teixeira, P.F., Berntsson, R.P., Murcha, M.W., Branca, R.M., Radomiljac, J.D., Regberg, J., Svensson, L.M., Bakali, A., Langel, U., Lehtio, J., Whelan, J., Stenmark, P., Glaser, E.(2013) Proc Natl Acad Sci U S A 110: E3761-E3769
- PubMed: 24043784
- DOI: https://doi.org/10.1073/pnas.1307637110
- Primary Citation of Related Structures:
4KA7, 4KA8 - PubMed Abstract:
Both mitochondria and chloroplasts contain distinct proteolytic systems for precursor protein processing catalyzed by the mitochondrial and stromal processing peptidases and for the degradation of targeting peptides catalyzed by presequence protease. Here, we have identified and characterized a component of the organellar proteolytic systems in Arabidopsis thaliana, the organellar oligopeptidase, OOP (At5g65620). OOP belongs to the M3A family of peptide-degrading metalloproteases. Using two independent in vivo methods, we show that the protease is dually localized to mitochondria and chloroplasts. Furthermore, we localized the OPP homolog At5g10540 to the cytosol. Analysis of peptide degradation by OOP revealed substrate size restriction from 8 to 23 aa residues. Short mitochondrial targeting peptides (presequence of the ribosomal protein L29 and presequence of 1-aminocyclopropane-1-carboxylic acid deaminase 1) and N- and C-terminal fragments derived from the presequence of the ATPase beta subunit ranging in size from 11 to 20 aa could be degraded. MS analysis showed that OOP does not exhibit a strict cleavage pattern but shows a weak preference for hydrophobic residues (F/L) at the P1 position. The crystal structures of OOP, at 1.8-1.9 Å, exhibit an ellipsoidal shape consisting of two major domains enclosing the catalytic cavity of 3,000 Å(3). The structural and biochemical data suggest that the protein undergoes conformational changes to allow peptide binding and proteolysis. Our results demonstrate the complementary role of OOP in targeting-peptide degradation in mitochondria and chloroplasts.
- Departments of Biochemistry and Biophysics and Neurochemistry, Arrhenius Laboratories for Natural Sciences, Stockholm University, SE-106 91 Stockholm, Sweden.
Organizational Affiliation: