Reshaping antibody diversity.
Wang, F., Ekiert, D.C., Ahmad, I., Yu, W., Zhang, Y., Bazirgan, O., Torkamani, A., Raudsepp, T., Mwangi, W., Criscitiello, M.F., Wilson, I.A., Schultz, P.G., Smider, V.V.(2013) Cell 153: 1379-1393
- PubMed: 23746848
- DOI: https://doi.org/10.1016/j.cell.2013.04.049
- Primary Citation of Related Structures:
4K3D, 4K3E - PubMed Abstract:
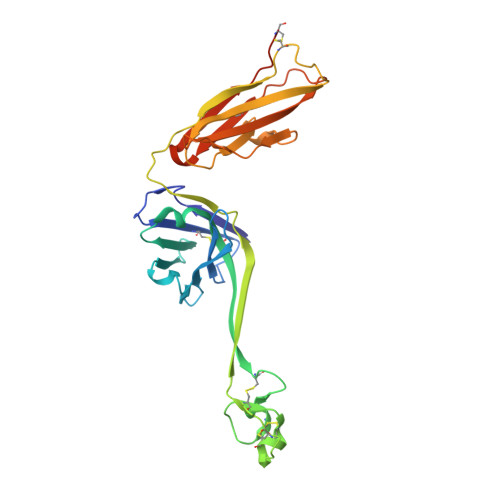
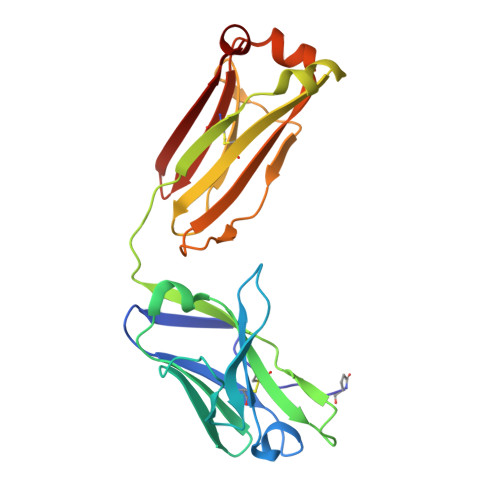
Some species mount a robust antibody response despite having limited genome-encoded combinatorial diversity potential. Cows are unusual in having exceptionally long CDR H3 loops and few V regions, but the mechanism for creating diversity is not understood. Deep sequencing reveals that ultralong CDR H3s contain a remarkable complexity of cysteines, suggesting that disulfide-bonded minidomains may arise during repertoire development. Indeed, crystal structures of two cow antibodies reveal that these CDR H3s form a very unusual architecture composed of a β strand "stalk" that supports a structurally diverse, disulfide-bonded "knob" domain. Diversity arises from somatic hypermutation of an ultralong DH with a severe codon bias toward mutation to cysteine. These unusual antibodies can be elicited to recognize defined antigens through the knob domain. Thus, the bovine immune system produces an antibody repertoire composed of ultralong CDR H3s that fold into a diversity of minidomains generated through combinations of somatically generated disulfides.
- Department of Chemistry, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: