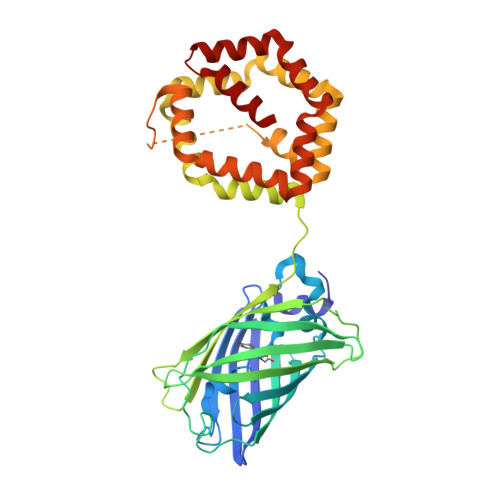
The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment.
Mueller, G.A., Pedersen, L.C., Lih, F.B., Glesner, J., Moon, A.F., Chapman, M.D., Tomer, K.B., London, R.E., Pomes, A.(2013) J Allergy Clin Immunol 132: 1420-1426.e9
- PubMed: 23915714
- DOI: https://doi.org/10.1016/j.jaci.2013.06.014
- Primary Citation of Related Structures:
4JRB - PubMed Abstract:
Sensitization to cockroach allergens is a major risk factor for asthma. The cockroach allergen Bla g 1 has multiple repeats of approximately 100 amino acids, but the fold of the protein and its biological function are unknown. We sought to determine the structure of Bla g 1, investigate the implications for allergic disease, and standardize cockroach exposure assays. nBla g 1 and recombinant constructs were compared by using ELISA with specific murine IgG and human IgE. The structure of Bla g 1 was determined by x-ray crystallography. Mass spectrometry and nuclear magnetic resonance spectroscopy were used to examine the ligand-binding properties of the allergen. The structure of an rBla g 1 construct with comparable IgE and IgG reactivity to the natural allergen was solved by x-ray crystallography. The Bla g 1 repeat forms a novel fold with 6 helices. Two repeats encapsulate a large and nearly spherical hydrophobic cavity, defining the basic structural unit. Lipids in the cavity varied depending on the allergen origin. Palmitic, oleic, and stearic acids were associated with nBla g 1 from cockroach frass. One unit of Bla g 1 was equivalent to 104 ng of allergen. Bla g 1 has a novel fold with a capacity to bind various lipids, which suggests a digestive function associated with nonspecific transport of lipid molecules in cockroaches. Defining the basic structural unit of Bla g 1 facilitates the standardization of assays in absolute units for the assessment of environmental allergen exposure.
- Laboratory of Structural Biology, National Institute of Environmental Health Sciences, Research Triangle Park, NC. Electronic address: Mueller3@niehs.nih.gov.
Organizational Affiliation: