Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
Bisht, S., Bharath, S.R., Murthy, M.R.N.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetylornithine/succinyldiaminopimelate aminotransferase | 422 | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | Mutation(s): 2 Gene Names: argD, dapC, dtu, STM3468 EC: 2.6.1.11 (PDB Primary Data), 2.6.1.17 (PDB Primary Data) | 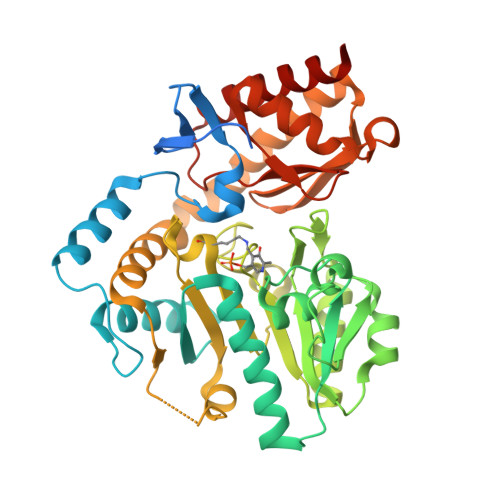 | |
UniProt | |||||
Find proteins for P40732 (Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)) Explore P40732 Go to UniProtKB: P40732 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P40732 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | C [auth A], D [auth A], G [auth B], J [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | E [auth A], I [auth B] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| NA Query on NA | F [auth A], H [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| LLP Query on LLP | A, B | L-PEPTIDE LINKING | C14 H22 N3 O7 P |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.914 | α = 90 |
| b = 113.604 | β = 90 |
| c = 69.183 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |