Crystal structure of wild type and D78N mutant Clavibacter michiganensis expansin, in apo form and in complex with oligosaccharides
Yennawar, N.H., Yennawar, H.P., Georgelis, N., Cosgrove, D.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cellulose binding protein | 202 | Clavibacter michiganensis subsp. michiganensis NCPPB 382 | Mutation(s): 0 Gene Names: celA, pCM1_0020 EC: 3.2.1.4 | 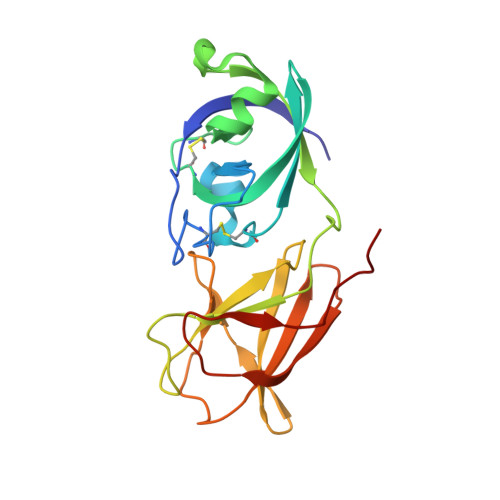 | |
UniProt | |||||
Find proteins for A5CLK3 (Clavibacter michiganensis subsp. michiganensis (strain NCPPB 382)) Explore A5CLK3 Go to UniProtKB: A5CLK3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5CLK3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900016 Query on PRD_900016 | C | beta-cellopentaose | Oligosaccharide / Metabolism |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 35.936 | α = 88.75 |
| b = 43.212 | β = 81.29 |
| c = 64.3 | γ = 82.7 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| PHENIX | model building |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |