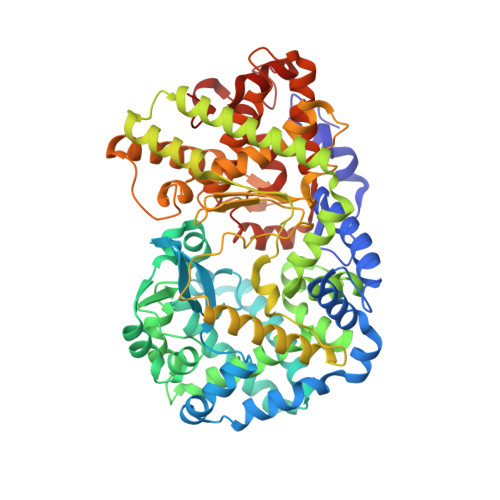
Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
Anderson, B.F., Knapp, K.M., Holland, R., Norris, G.E., Christensson, C., Bratt, H., Collins, L.J., Coolbear, T., Lubbers, M.W., O Toole, P.W., Reid, J.R., Jameson, G.B.To be published.