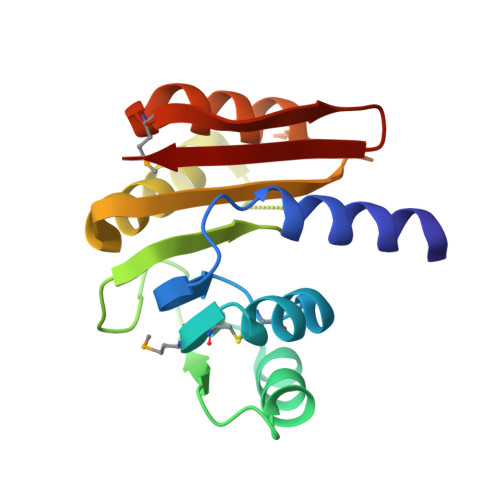
Crystal structure of a putative Methyltransferase from Pseudomonas syringae
Filippova, E.V., Wawrzak, Z., Minasov, G., Shuvalova, L., Kiryukhina, O., Clancy, S., Joachimiak, A., Anderson, W.F., Midwest Center for Structural Genomics (MCSG)To be published.