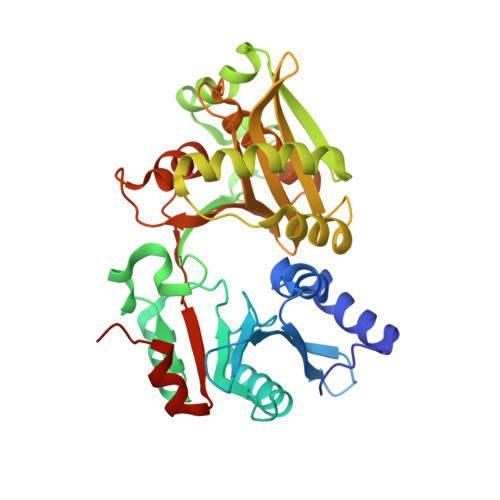
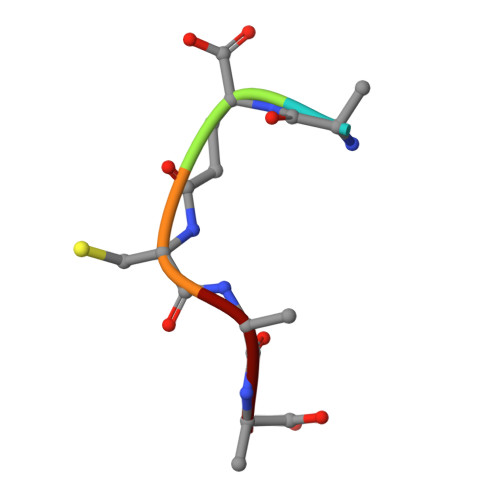
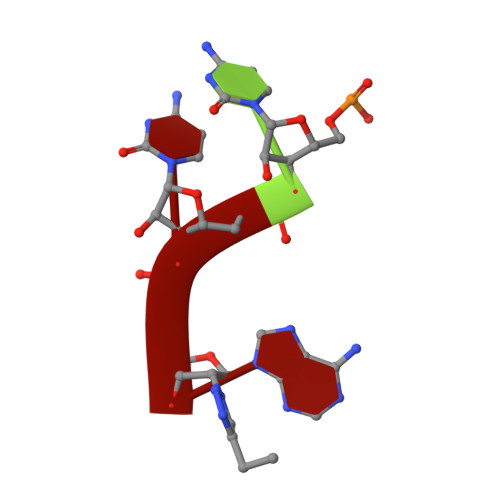
The Structure of FemXWv in Complex with a Peptidyl-RNA Conjugate: Mechanism of Aminoacyl Transfer from Ala-tRNA(Ala) to Peptidoglycan Precursors
Fonvielle, M., Li de La Sierra-Gallay, I., El-Sagheer, A.H., Lecerf, M., Patin, D., Mellal, D., Mayer, C., Blanot, D., Gale, N., Brown, T., van Tilbeurgh, H., Etheve-Quelquejeu, M., Arthur, M.(2013) Angew Chem Int Ed Engl 52: 7278-7281
- PubMed: 23744707
- DOI: https://doi.org/10.1002/anie.201301411
- Primary Citation of Related Structures:
4II9 - Laboratoire de Recherche Moléculaire sur les Antibiotiques, Centre de Recherche des Cordeliers, Equipe 12, INSERM, U872, 75006 Paris, France.
Organizational Affiliation: