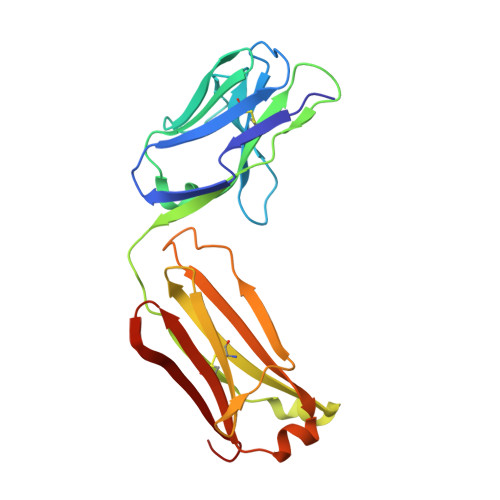
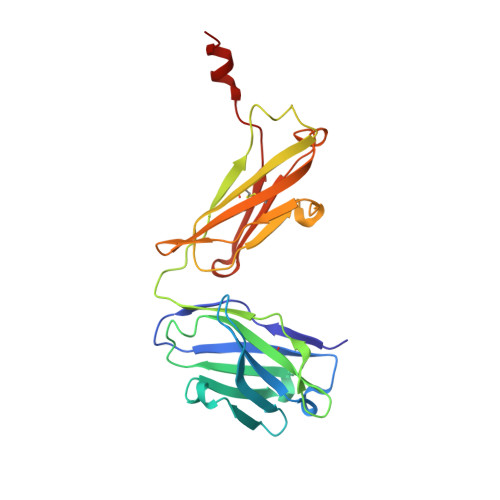
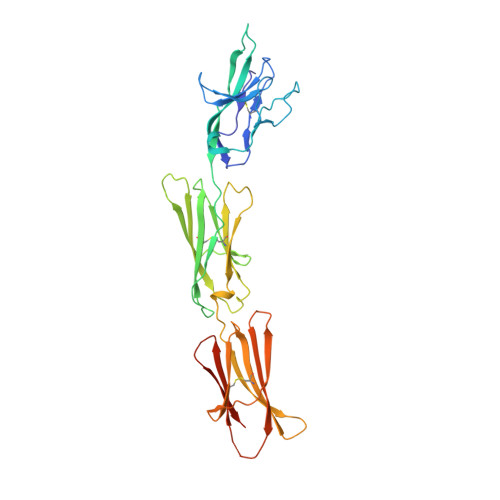
Crystal structure of signal regulatory protein gamma (SIRP gamma) in complex with an antibody Fab fragment.
Nettleship, J.E., Ren, J., Scott, D.J., Rahman, N., Hatherley, D., Zhao, Y., Stuart, D.I., Barclay, A.N., Owens, R.J.(2013) BMC Struct Biol 13: 13-13
- PubMed: 23826770
- DOI: https://doi.org/10.1186/1472-6807-13-13
- Primary Citation of Related Structures:
4I2X - PubMed Abstract:
Signal Regulatory Protein γ (SIRPγ) is a member of a closely related family of three cell surface receptors implicated in modulating immune/inflammatory responses. SIRPγ is expressed on T lymphocytes where it appears to be involved in the integrin-independent adhesion of lymphocytes to antigen-presenting cells. Here we describe the first full length structure of the extracellular region of human SIRPγ. We obtained crystals of SIRPγ by making a complex of the protein with the Fab fragment of the anti-SIRP antibody, OX117, which also binds to SIRPα and SIRPβ. We show that the epitope for FabOX117 is formed at the interface of the first and second domains of SIRPγ and comprises residues which are conserved between all three SIRPs. The FabOX117 binding site is distinct from the region in domain 1 which interacts with CD47, the physiological ligand for both SIRPγ and SIRPα but not SIRPβ. Comparison of the three domain structures of SIRPγ and SIRPα showed that these receptors can adopt different overall conformations due to the flexibility of the linker between the first two domains. SIRPγ in complex with FabOX117 forms a dimer in the crystal. Binding to the Fab fixes the position of domain 1 relative to domains 2/3 exposing a surface which favours formation of a homotypic dimer. However, the interaction appears to be relatively weak since only monomers of SIRPγ were observed in sedimentation velocity analytical ultracentrifugation of the protein alone. Studies of complex formation by equilibrium ultracentrifugation showed that only a 1:1 complex of SIRPγ: FabOX117 was formed with a dissociation constant in the low micromolar range (Kd = 1.2 +/- 0.3 μM). The three-domain extracellular regions of SIRPs are structurally conserved but show conformational flexibility in the disposition of the amino terminal ligand-binding Ig domain relative to the two membrane proximal Ig domains. Binding of a cross-reactive anti-SIRP Fab fragment to SIRPγ stabilises a conformation that favours SIRP dimer formation in the crystal structure, though this interaction does not appear sufficiently stable to be observed in solution.