1.2 structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain collected at wavelength 0.91974
Stiegler, A.L., Jakoncic, J., Stojanoff, V., Chiswell, B.P., Calderwood, D.A., Boggon, T.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Integrin-linked protein kinase | 179 | Homo sapiens | Mutation(s): 0 Gene Names: ILK, ILK1, ILK2 EC: 2.7.11.1 | 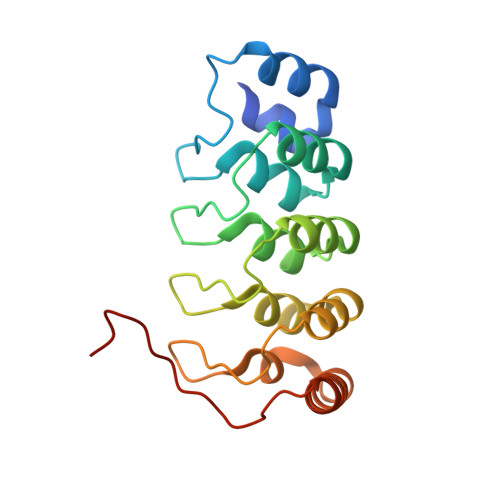 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13418 (Homo sapiens) Explore Q13418 Go to UniProtKB: Q13418 | |||||
PHAROS: Q13418 GTEx: ENSG00000166333 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13418 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LIM and senescent cell antigen-like-containing domain protein 1 | 72 | Homo sapiens | Mutation(s): 0 Gene Names: LIMS1, PINCH, PINCH1 | 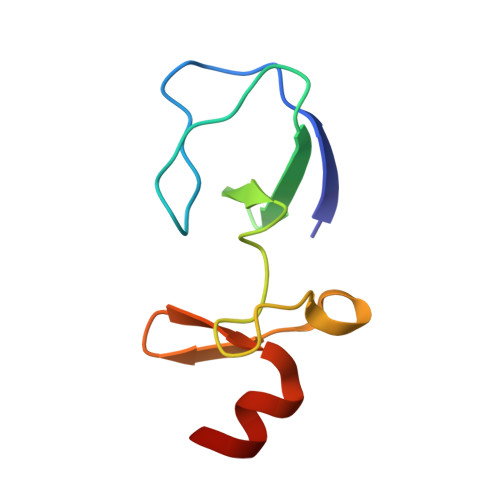 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P48059 (Homo sapiens) Explore P48059 Go to UniProtKB: P48059 | |||||
PHAROS: P48059 GTEx: ENSG00000169756 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P48059 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| IOD Query on IOD | D [auth A] E [auth A] F [auth A] G [auth A] H [auth A] | IODIDE ION I XMBWDFGMSWQBCA-UHFFFAOYSA-M |  | ||
| PO4 Query on PO4 | C [auth A] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| ZN Query on ZN | L [auth B], M [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 37.144 | α = 78.67 |
| b = 41.775 | β = 68.94 |
| c = 46.798 | γ = 85.81 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| Executor | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |