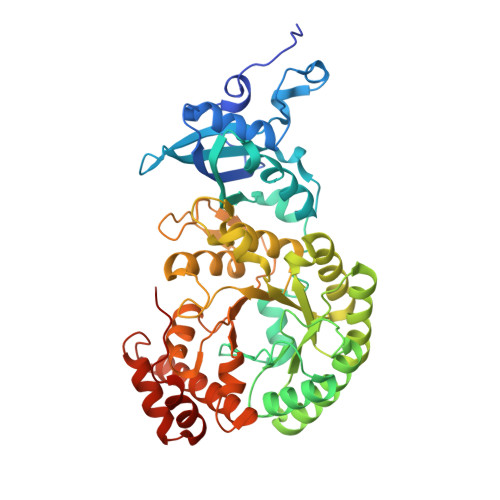
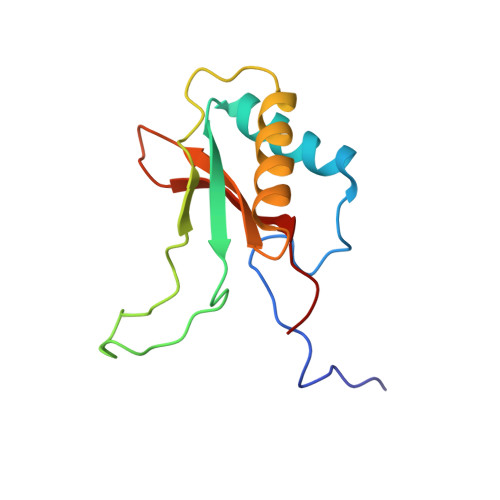
Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate.
Loewen, P.C., Didychuk, A.L., Switala, J., Perez-Luque, R., Fita, I., Loewen, M.C.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 10-14
- PubMed: 23295478
- DOI: https://doi.org/10.1107/S1744309112047549
- Primary Citation of Related Structures:
4HHH - PubMed Abstract:
The first structure of a ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from a pulse crop is reported. Rubisco was purified from Pisum sativum (garden pea) and diffraction-quality crystals were obtained by hanging-drop vapour diffusion in the presence of the substrate ribulose 1,5-bisphosphate. X-ray diffraction data were recorded to 2.20 Å resolution from a single crystal at the Canadian Light Source. The overall quaternary structure of non-activated P. sativum Rubisco highlights the conservation of the form I Rubisco hexadecameric complex. The electron density places the substrate in the active site at the interface of the large-subunit dimers. Lys201 in the active site is not carbamylated as expected for this non-activated structure. Some heterogeneity in the small-subunit sequence is noted, as well as possible variations in the conformation and contacts of ribulose 1,5-bisphosphate in the large-subunit active sites. Overall, the active-site conformation most closely correlates with the `closed' conformation observed in other substrate/inhibitor-bound Rubisco structures.
- Department of Microbiology, University of Manitoba, 418 Buller Building, Winnipeg, MB R3T 2N2, Canada.
Organizational Affiliation: