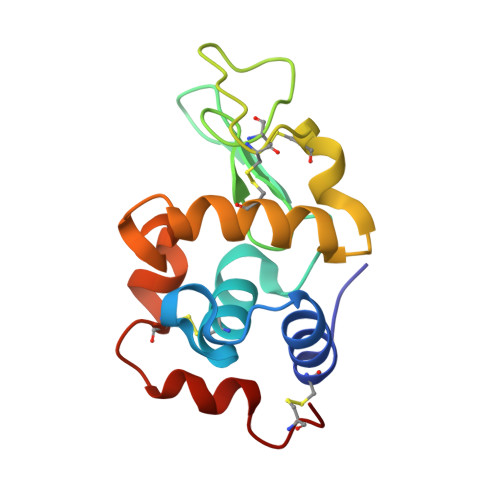
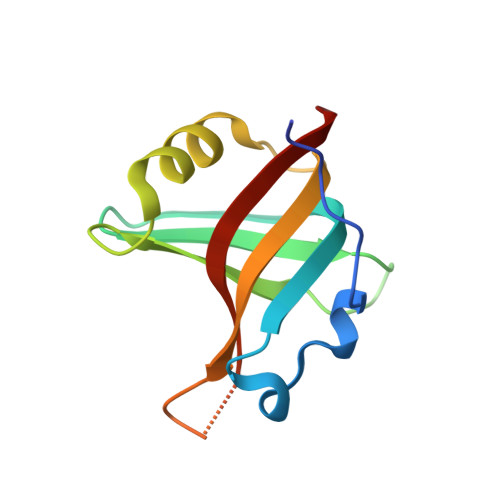
Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Steemson, J.D., Baake, M., Rakonjac, J., Arcus, V.L., Liddament, M.T.(2014) PLoS One 9: e86050-e86050
- PubMed: 24465865
- DOI: https://doi.org/10.1371/journal.pone.0086050
- Primary Citation of Related Structures:
4GLA, 4GLV, 4GN3, 4GN4, 4GN5 - PubMed Abstract:
The OB-fold is a small, versatile single-domain protein binding module that occurs in all forms of life, where it binds protein, carbohydrate, nucleic acid and small-molecule ligands. We have exploited this natural plasticity to engineer a new class of non-immunoglobulin alternatives to antibodies with unique structural and biophysical characteristics. We present here the engineering of the OB-fold anticodon recognition domain from aspartyl tRNA synthetase taken from the thermophile Pyrobaculum aerophilum. For this single-domain scaffold we have coined the term OBody. Starting from a naïve combinatorial library, we engineered an OBody with 3 nM affinity for hen egg-white lysozyme, by optimising the affinity of a naïve OBody 11,700-fold over several affinity maturation steps, using phage display. At each maturation step a crystal structure of the engineered OBody in complex with hen egg-white lysozyme was determined, showing binding elements in atomic detail. These structures have given us an unprecedented insight into the directed evolution of affinity for a single antigen on the molecular scale. The engineered OBodies retain the high thermal stability of the parental OB-fold despite mutation of up to 22% of their residues. They can be expressed in soluble form and also purified from bacteria at high yields. They also lack disulfide bonds. These data demonstrate the potential of OBodies as a new scaffold for the engineering of specific binding reagents and provide a platform for further development of future OBody-based applications.
- Department of Biological Sciences, University of Waikato, Hamilton, New Zealand.
Organizational Affiliation: