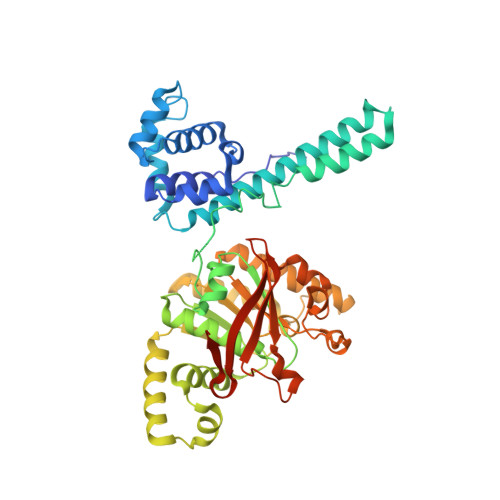
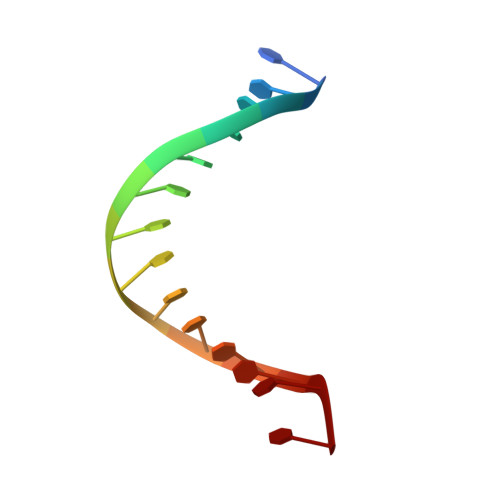
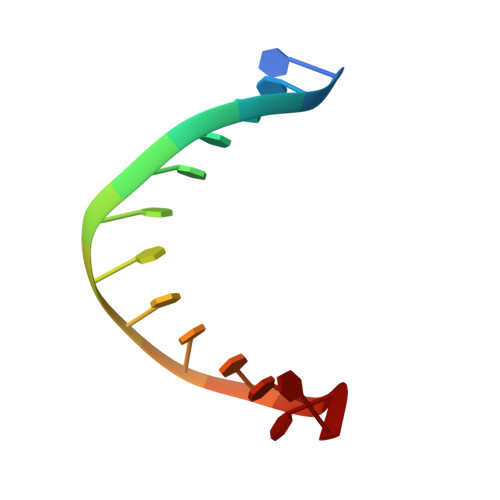
The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation.
Itsathitphaisarn, O., Wing, R.A., Eliason, W.K., Wang, J., Steitz, T.A.(2012) Cell 151: 267-277
- PubMed: 23022319
- DOI: https://doi.org/10.1016/j.cell.2012.09.014
- Primary Citation of Related Structures:
4ESV - PubMed Abstract:
DNA polymerases can only synthesize nascent DNA from single-stranded DNA (ssDNA) templates. In bacteria, the unwinding of parental duplex DNA is carried out by the replicative DNA helicase (DnaB) that couples NTP hydrolysis to 5' to 3' translocation. The crystal structure of the DnaB hexamer in complex with GDP-AlF(4) and ssDNA reported here reveals that DnaB adopts a closed spiral staircase quaternary structure around an A-form ssDNA with each C-terminal domain coordinating two nucleotides of ssDNA. The structure not only provides structural insights into the translocation mechanism of superfamily IV helicases but also suggests that members of this superfamily employ a translocation mechanism that is distinct from other helicase superfamilies. We propose a hand-over-hand mechanism in which sequential hydrolysis of NTP causes a sequential 5' to 3' movement of the subunits along the helical axis of the staircase, resulting in the unwinding of two nucleotides per subunit.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: