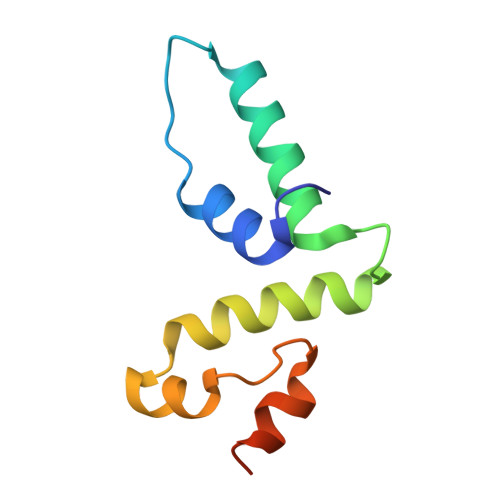
Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206.
Liang, Y., Choo, S.H., Rossbach, M., Baburajendran, N., Palasingam, P., Kolatkar, P.R.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 443-447
- PubMed: 22505416
- DOI: https://doi.org/10.1107/S1744309112006070
- Primary Citation of Related Structures:
4E6S - PubMed Abstract:
Zfp206 (also named Zscan10) is a transcription factor that plays an important role in maintaining the pluripotent state of embryonic stem cells. Zfp206 is a member of the SCAN-domain family of C(2)H(2) zinc-finger transcription factors. The SCAN domain is a highly conserved motif of 84 residues which mediates the self-association of and heterodimerization between SCAN-domain family transcription factors. The SCAN domain may therefore be the key to the selective oligomerization of and may combinatorially enhance the regulatory versatility of C(2)H(2) zinc fingers. This paper describes crystallization attempts with the SCAN domain of Zfp206 (Zfp206SCAN) and optimization strategies to obtain diffraction-quality crystals. The best diffracting crystal was grown in a solution consisting of 0.3 M ammonium sulfate, 0.1 M Tris-HCl pH 8.6, 25% PEG 3350, 0.1 M ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA) using the hanging-drop vapour-diffusion technique. Optimized crystals diffracted to 1.85 Å resolution and belonged to space group I422, with unit-cell parameters a = 67.57, c = 87.54 Å. A Matthews analysis indicated the presence of one Zfp206SCAN molecule per asymmetric unit.
- Laboratory for Structural Biochemistry, Genome Institute of Singapore, Genome, 60 Biopolis Street, Singapore 138672, Singapore.
Organizational Affiliation: