Structural and Biochemical Analyses of a Clostridium Perfringens Sortase D Transpeptidase
Suryadinata, R., Seabrook, S.A., Adams, T.E., Nuttall, S.D., Peat, T.S.(2015) Acta Crystallogr D Biol Crystallogr 71: 1505
- PubMed: 26143922
- DOI: https://doi.org/10.1107/S1399004715009219
- Primary Citation of Related Structures:
4D70 - PubMed Abstract:
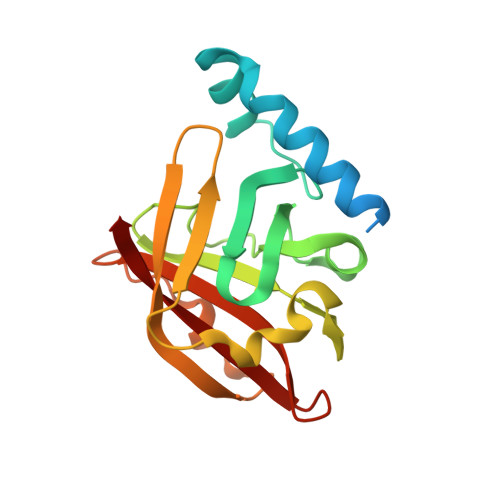
The assembly and anchorage of various pathogenic proteins on the surface of Gram-positive bacteria is mediated by the sortase family of enzymes. These cysteine transpeptidases catalyze a unique sorting signal motif located at the C-terminus of their target substrate and promote the covalent attachment of these proteins onto an amino nucleophile located on another protein or on the bacterial cell wall. Each of the six distinct classes of sortases displays a unique biological role, with sequential activation of multiple sortases often observed in many Gram-positive bacteria to decorate their peptidoglycans. Less is known about the members of the class D family of sortases (SrtD), but they have a suggested role in spore formation in an oxygen-limiting environment. Here, the crystal structure of the SrtD enzyme from Clostridium perfringens was determined at 1.99 Å resolution. Comparative analysis of the C. perfringens SrtD structure reveals the typical eight-stranded β-barrel fold observed in all other known sortases, along with the conserved catalytic triad consisting of cysteine, histidine and arginine residues. Biochemical approaches further reveal the specifics of the SrtD catalytic activity in vitro, with a significant preference for the LPQTGS sorting motif. Additionally, the catalytic activity of SrtD is most efficient at 316 K and can be further improved in the presence of magnesium cations. Since C. perfringens spores are heat-resistant and lead to foodborne illnesses, characterization of the spore-promoting sortase SrtD may lead to the development of new antimicrobial agents.
- Manufacturing Flagship, Commonwealth Scientific and Industrial Research Organisation, 343 Royal Parade, Parkville, Victoria 3052, Australia.
Organizational Affiliation: