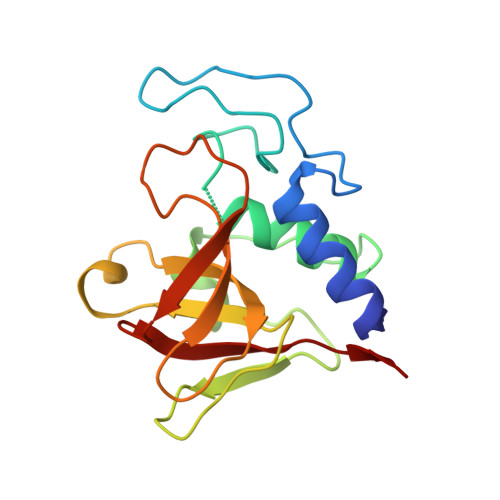
Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Sanz-Gaitero, M., Keary, R., Garcia-Doval, C., Coffey, A., van Raaij, M.J.(2014) Virol J 11: 133-133
- PubMed: 25064136
- DOI: https://doi.org/10.1186/1743-422X-11-133
- Primary Citation of Related Structures:
4CSH, 4CT3 - PubMed Abstract:
Bacteriophages encode endolysins to lyse their host cell and allow escape of their progeny. Endolysins are also active against Gram-positive bacteria when applied from the outside and are thus attractive anti-bacterial agents. LysK, an endolysin from staphylococcal phage K, contains an N-terminal cysteine-histidine dependent amido-hydrolase/peptidase domain (CHAP(K)), a central amidase domain and a C-terminal SH3b cell wall-binding domain. CHAP(K) cleaves bacterial peptidoglycan between the tetra-peptide stem and the penta-glycine bridge. The CHAP(K) domain of LysK was crystallized and high-resolution diffraction data was collected both from a native protein crystal and a methylmercury chloride derivatized crystal. The anomalous signal contained in the derivative data allowed the location of heavy atom sites and phase determination. The resulting structures were completed, refined and analyzed. The presence of calcium and zinc ions in the structure was confirmed by X-ray fluorescence emission spectroscopy. Zymogram analysis was performed on the enzyme and selected site-directed mutants. The structure of CHAP(K) revealed a papain-like topology with a hydrophobic cleft, where the catalytic triad is located. Ordered buffer molecules present in this groove may mimic the peptidoglycan substrate. When compared to previously solved CHAP domains, CHAP(K) contains an additional lobe in its N-terminal domain, with a structural calcium ion, coordinated by residues Asp45, Asp47, Tyr49, His51 and Asp56. The presence of a zinc ion in the active site was also apparent, coordinated by the catalytic residue Cys54 and a possible substrate analogue. Site-directed mutagenesis was used to demonstrate that residues involved in calcium binding and of the proposed active site were important for enzyme activity. The high-resolution structure of the CHAP(K) domain of LysK was determined, suggesting the location of the active site, the substrate-binding groove and revealing the presence of a structurally important calcium ion. A zinc ion was found more loosely bound. Based on the structure, we propose a possible reaction mechanism. Future studies will be aimed at co-crystallizing CHAP(K) with substrate analogues and elucidating its role in the complete LysK protein. This, in turn, may lead to the design of site-directed mutants with altered activity or substrate specificity.
- Departamento de Estructura de Macromoleculas, Centro Nacional de Biotecnologia (CNB-CSIC), Calle Darwin 3, E-28049 Madrid, Spain. mjvanraaij@cnb.csic.es.
Organizational Affiliation: