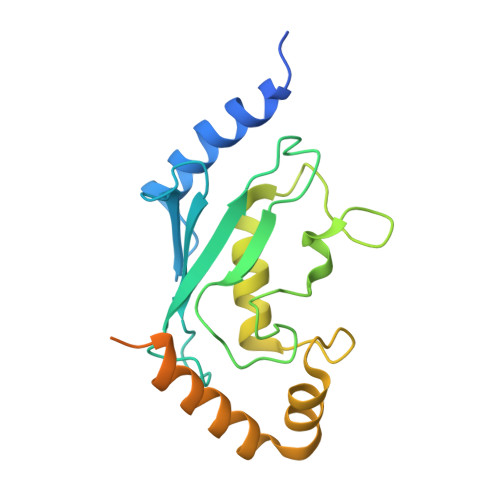
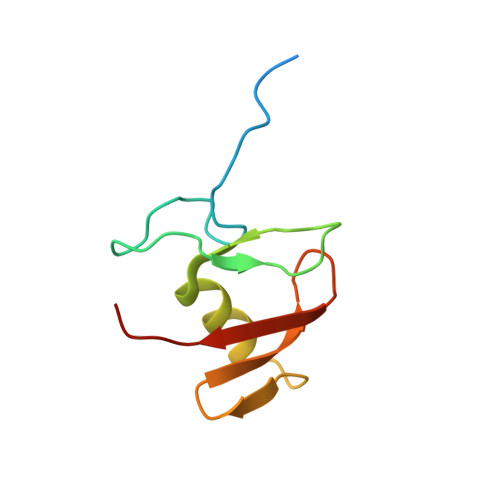
Structure of the Human Fancl Ring-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection.
Hodson, C., Purkiss, A., Miles, J.A., Walden, H.(2014) Structure 22: 337
- PubMed: 24389026
- DOI: https://doi.org/10.1016/j.str.2013.12.004
- Primary Citation of Related Structures:
4CCG - PubMed Abstract:
The combination of an E2 ubiquitin-conjugating enzyme with an E3 ubiquitin-ligase is essential for ubiquitin modification of a substrate. Moreover, the pairing dictates both the substrate choice and the modification type. The molecular details of generic E3-E2 interactions are well established. Nevertheless, the determinants of selective, specific E3-E2 recognition are not understood. There are ∼40 E2s and ∼600 E3s giving rise to a possible ∼24,000 E3-E2 pairs. Using the Fanconi Anemia pathway exclusive E3-E2 pair, FANCL-Ube2T, we report the atomic structure of the FANCL RING-Ube2T complex, revealing a specific and extensive network of additional electrostatic and hydrophobic interactions. Furthermore, we show that these specific interactions are required for selection of Ube2T over other E2s by FANCL.
- Protein Structure and Function Laboratory, Lincoln's Inn Fields Laboratories of the London Research Institute, Cancer Research UK, 44 Lincoln's Inn Fields, London WC2A 3LY, UK.
Organizational Affiliation: