Insight Into Structural Evolution of Extremophilic Proteins
Talon, R., Girard, E., Franzetti, B., Madern, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MALATE DEHYDROGENASE | 323 | Picrophilus oshimae | Mutation(s): 0 EC: 1.1.1.37 | 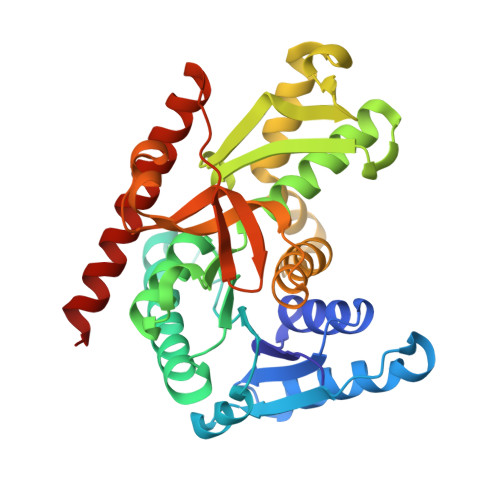 | |
UniProt | |||||
Find proteins for Q6L0C3 (Picrophilus torridus (strain ATCC 700027 / DSM 9790 / JCM 10055 / NBRC 100828 / KAW 2/3)) Explore Q6L0C3 Go to UniProtKB: Q6L0C3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6L0C3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PG6 Query on PG6 | L [auth C] | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE C12 H26 O6 DMDPGPKXQDIQQG-UHFFFAOYSA-N |  | ||
| CIT Query on CIT | G [auth B] | CITRIC ACID C6 H8 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | E [auth A] F [auth A] H [auth B] I [auth B] J [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.167 | α = 90 |
| b = 81.167 | β = 90 |
| c = 395.99 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| PHENIX | refinement |