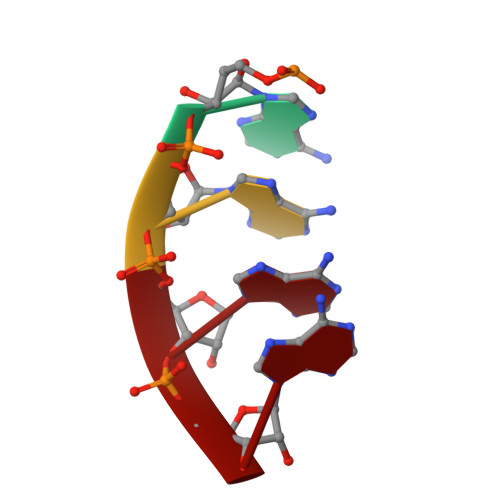
Crystal Structure of a 9-Subunit Archaeal Exosome in Pre-Catalytic States of the Phosphorolytic Reaction.
Lorentzen, E., Conti, E.(2012) Archaea 2012: 21869
- PubMed: 23319881
- DOI: https://doi.org/10.1155/2012/721869
- Primary Citation of Related Structures:
4BA1, 4BA2 - PubMed Abstract:
The RNA exosome is an important protein complex that functions in the 3' processing and degradation of RNA in archaeal and eukaryotic organisms. The archaeal exosome is functionally similar to bacterial polynucleotide phosphorylase (PNPase) and RNase PH enzymes as it uses inorganic phosphate (Pi) to processively cleave RNA substrates releasing nucleoside diphosphates. To shed light on the mechanism of catalysis, we have determined the crystal structures of mutant archaeal exosome in complex with either Pi or with both RNA and Pi at resolutions of 1.8 Å and 2.5 Å, respectively. These structures represent views of precatalytic states of the enzyme and allow the accurate determination of the substrate binding geometries. In the structure with both Pi and RNA bound, the Pi closely approaches the phosphate of the 3'-end nucleotide of the RNA and is in a perfect position to perform a nucleophilic attack. The presence of negative charge resulting from the close contacts between the phosphates appears to be neutralized by conserved positively charged residues in the active site of the archaeal exosome. The high degree of structural conservation between the archaeal exosome and the PNPase including the requirement for divalent metal ions for catalysis is discussed.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany.
Organizational Affiliation: