Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Mariotti, P., Malito, E., Biancucci, M., Lo Surdo, P., Mishra, R.P., Nardi-Dei, V., Savino, S., Nissum, M., Spraggon, G., Grandi, G., Bagnoli, F., Bottomley, M.J.(2013) Biochem J 449: 683
- PubMed: 23113737
- DOI: https://doi.org/10.1042/BJ20121426
- Primary Citation of Related Structures:
4B8Y - PubMed Abstract:
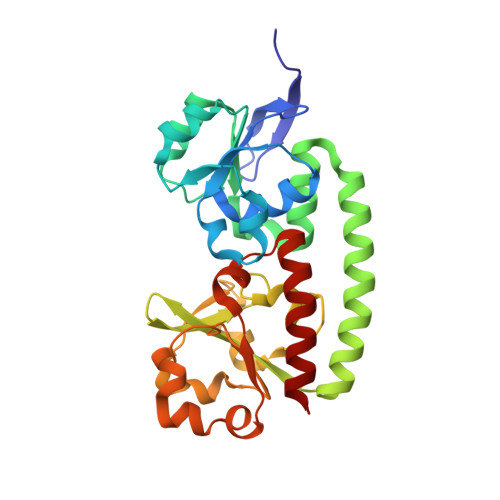
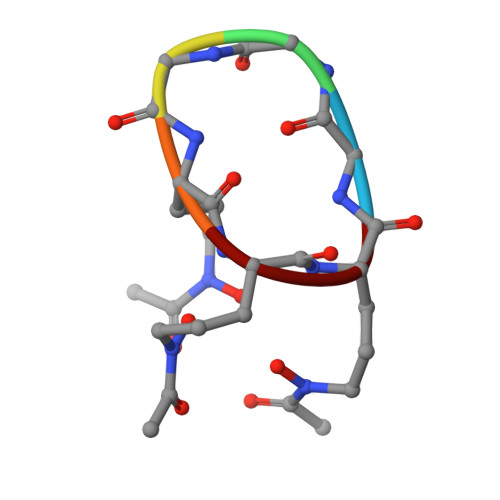
Staphylococcus aureus is a human pathogen causing globally significant morbidity and mortality. The development of antibiotic resistance in S. aureus highlights the need for a preventive vaccine. In the present paper we explore the structure and function of FhuD2 (ferric-hydroxamate uptake D2), a staphylococcal surface lipoprotein mediating iron uptake during invasive infection, recently described as a promising vaccine candidate. Differential scanning fluorimetry and calorimetry studies revealed that FhuD2 is stabilized by hydroxamate siderophores. The FhuD2-ferrichrome interaction was of nanomolar affinity in surface plasmon resonance experiments and fully iron(III)-dependent. We determined the X-ray crystallographic structure of ligand-bound FhuD2 at 1.9 Å (1 Å=0.1 nm) resolution, revealing the bilobate fold of class III SBPs (solute-binding proteins). The ligand, ferrichrome, occupies a cleft between the FhuD2 N- and C-terminal lobes. Many FhuD2-siderophore interactions enable the specific recognition of ferrichrome. Biochemical data suggest that FhuD2 does not undergo significant conformational changes upon siderophore binding, supporting the hypothesis that the ligand-bound complex is essential for receptor engagement and uptake. Finally, immunizations with FhuD2 alone or FhuD2 formulated with hydroxamate siderophores were equally protective in a murine staphylococcal infection model, confirming the suitability and efficacy of apo-FhuD2 as a protective antigen, and suggesting that other class III SBPs might also be exploited as vaccine candidates.
- Novartis Vaccines and Diagnostics srl, Via Fiorentina 1, 53100 Siena, Italy.
Organizational Affiliation: