Sequence-Specific Transcription Factor NF-Y Displays Histone-Like DNA Binding and H2B-Like Ubiquitination.
Nardini, M., Gnesutta, N., Donati, G., Gatta, R., Forni, C., Fossati, A., Vonrhein, C., Moras, D., Romier, C., Bolognesi, M., Mantovani, R.(2013) Cell 152: 132
- PubMed: 23332751
- DOI: https://doi.org/10.1016/j.cell.2012.11.047
- Primary Citation of Related Structures:
4AWL - PubMed Abstract:
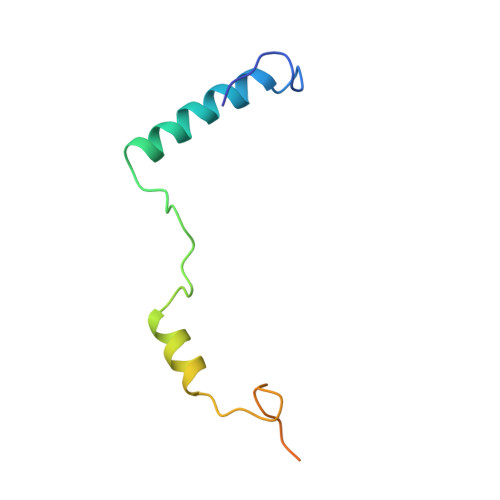
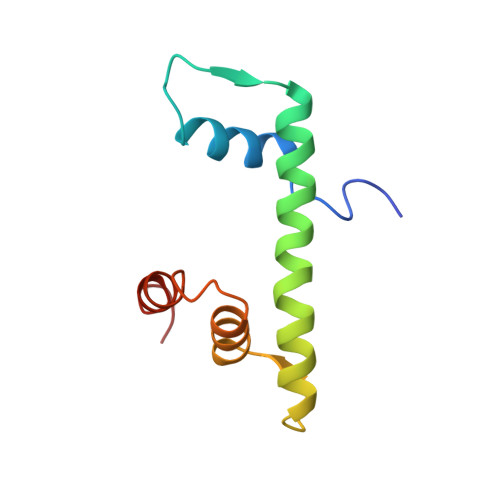
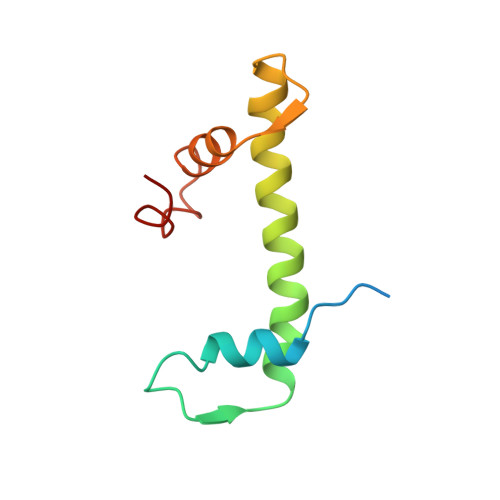
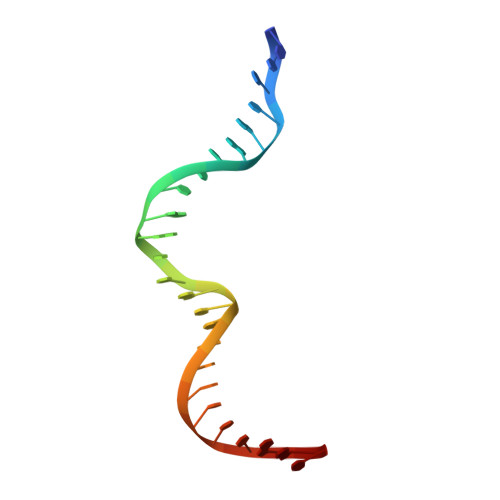
The sequence-specific transcription factor NF-Y binds the CCAAT box, one of the sequence elements most frequently found in eukaryotic promoters. NF-Y is composed of the NF-YA and NF-YB/NF-YC subunits, the latter two hosting histone-fold domains (HFDs). The crystal structure of NF-Y bound to a 25 bp CCAAT oligonucleotide shows that the HFD dimer binds to the DNA sugar-phosphate backbone, mimicking the nucleosome H2A/H2B-DNA assembly. NF-YA both binds to NF-YB/NF-YC and inserts an α helix deeply into the DNA minor groove, providing sequence-specific contacts to the CCAAT box. Structural considerations and mutational data indicate that NF-YB ubiquitination at Lys138 precedes and is equivalent to H2B Lys120 monoubiquitination, important in transcriptional activation. Thus, NF-Y is a sequence-specific transcription factor with nucleosome-like properties of nonspecific DNA binding and helps establish permissive chromatin modifications at CCAAT promoters. Our findings suggest that other HFD-containing proteins may function in similar ways.
- Dipartimento di BioScienze, Università degli Studi di Milano, Via Celoria 26, 20133 Milano, Italy.
Organizational Affiliation: