Structure and Assembly of a Trans-Periplasmic Channel for Type Iv Pili in Neisseria Meningitidis.
Berry, J.L., Phelan, M.M., Collins, R.F., Adomavicius, T., Tonjum, T., Frye, S.A., Bird, L., Owens, R., Ford, R.C., Lian, L.Y., Derrick, J.P.(2012) PLoS Pathog 8: 2923
- PubMed: 23028322
- DOI: https://doi.org/10.1371/journal.ppat.1002923
- Primary Citation of Related Structures:
4AQZ, 4AR0, 4AV2 - PubMed Abstract:
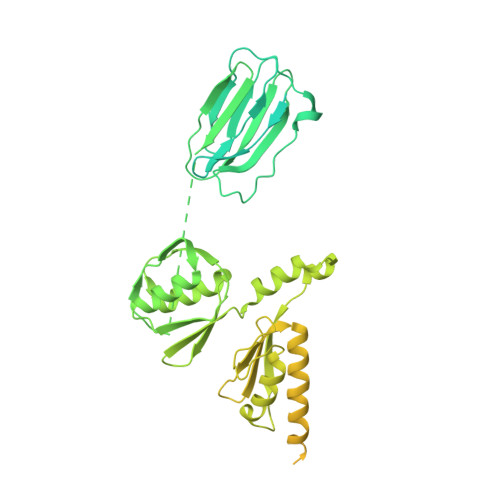
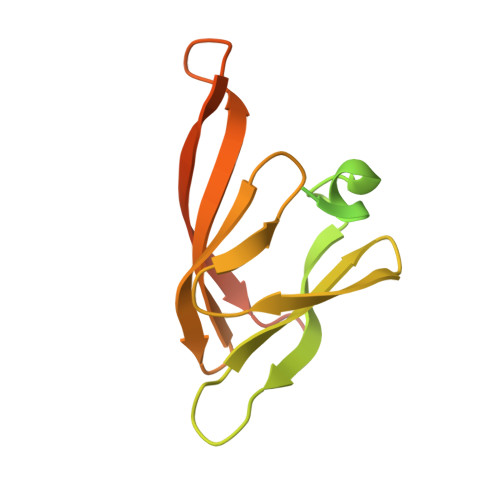
Type IV pili are polymeric fibers which protrude from the cell surface and play a critical role in adhesion and invasion by pathogenic bacteria. The secretion of pili across the periplasm and outer membrane is mediated by a specialized secretin protein, PilQ, but the way in which this large channel is formed is unknown. Using NMR, we derived the structures of the periplasmic domains from N. meningitidis PilQ: the N-terminus is shown to consist of two β-domains, which are unique to the type IV pilus-dependent secretins. The structure of the second β-domain revealed an eight-stranded β-sandwich structure which is a novel variant of the HSP20-like fold. The central part of PilQ consists of two α/β fold domains: the structure of the first of these is similar to domains from other secretins, but with an additional α-helix which links it to the second α/β domain. We also determined the structure of the entire PilQ dodecamer by cryoelectron microscopy: it forms a cage-like structure, enclosing a cavity which is approximately 55 Å in internal diameter at its largest extent. Specific regions were identified in the density map which corresponded to the individual PilQ domains: this allowed us to dock them into the cryoelectron microscopy density map, and hence reconstruct the entire PilQ assembly which spans the periplasm. We also show that the C-terminal domain from the lipoprotein PilP, which is essential for pilus assembly, binds specifically to the first α/β domain in PilQ and use NMR chemical shift mapping to generate a model for the PilP:PilQ complex. We conclude that passage of the pilus fiber requires disassembly of both the membrane-spanning and the β-domain regions in PilQ, and that PilP plays an important role in stabilising the PilQ assembly during secretion, through its anchorage in the inner membrane.
- Faculty of Life Sciences, Michael Smith Building, University of Manchester, Manchester, United Kingdom.
Organizational Affiliation: