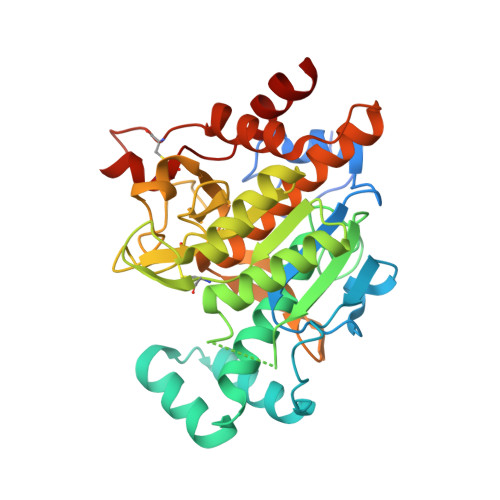
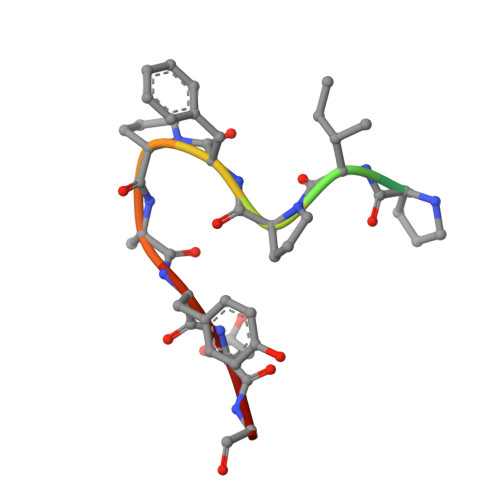
The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Koehnke, J., Bent, A., Houssen, W.E., Zollman, D., Morawitz, F., Shirran, S., Vendome, J., Nneoyiegbe, A.F., Trembleau, L., Botting, C.H., Smith, M.C., Jaspars, M., Naismith, J.H.(2012) Nat Struct Mol Biol 19: 767
- PubMed: 22796963
- DOI: https://doi.org/10.1038/nsmb.2340
- Primary Citation of Related Structures:
4AKS, 4AKT - PubMed Abstract:
Peptide macrocycles are found in many biologically active natural products. Their versatility, resistance to proteolysis and ability to traverse membranes has made them desirable molecules. Although technologies exist to synthesize such compounds, the full extent of diversity found among natural macrocycles has yet to be achieved synthetically. Cyanobactins are ribosomal peptide macrocycles encompassing an extraordinarily diverse range of ring sizes, amino acids and chemical modifications. We report the structure, biochemical characterization and initial engineering of the PatG macrocyclase domain of Prochloron sp. from the patellamide pathway that catalyzes the macrocyclization of linear peptides. The enzyme contains insertions in the subtilisin fold to allow it to recognize a three-residue signature, bind substrate in a preorganized and unusual conformation, shield an acyl-enzyme intermediate from water and catalyze peptide bond formation. The ability to macrocyclize a broad range of nonactivated substrates has wide biotechnology applications.
- Biomedical Sciences Research Complex, University of St Andrews, St Andrews, UK.
Organizational Affiliation: